4PB1
 
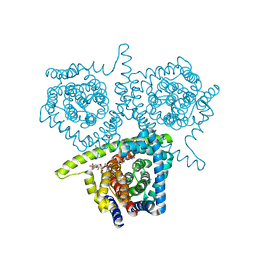 | | Structure of vcCNT-7C8C bound to ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD9
 
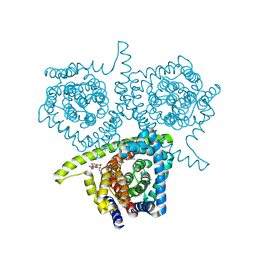 | | Structure of vcCNT-7C8C bound to adenosine | | Descriptor: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
3TD5
 
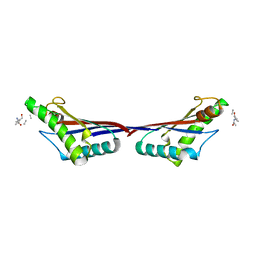 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala | | Descriptor: | CHLORIDE ION, Outer membrane protein omp38, peptide(L-Ala-gamma-D-Glu-m-DAP-D-Ala-D-Ala) | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD4
 
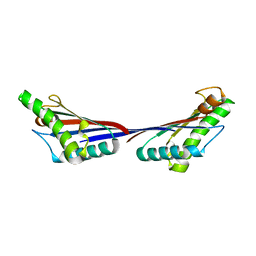 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
3TD3
 
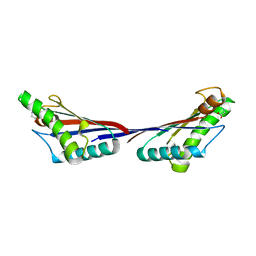 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
5KUC
 
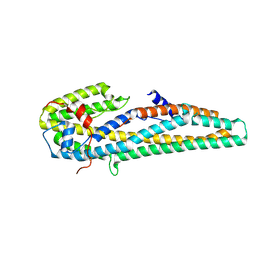 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
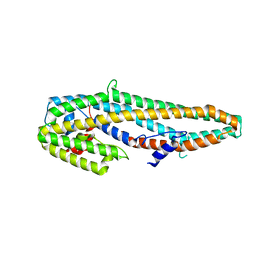 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
8G4T
 
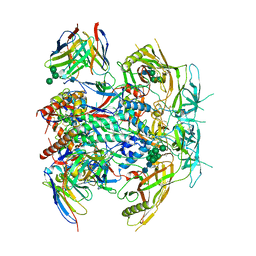 | | Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein BG505 DS-SOSIP gp120, ... | | Authors: | Wang, S, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
8G4M
 
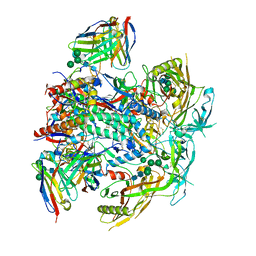 | | Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Wang, S, Morano, N.C, Shapiro, L, Kwong, P.D. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | HIV-1 neutralizing antibodies elicited in humans by a prefusion-stabilized envelope trimer form a reproducible class targeting fusion peptide.
Cell Rep, 42, 2023
|
|
7PL3
 
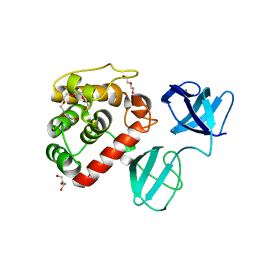 | |
7PJ6
 
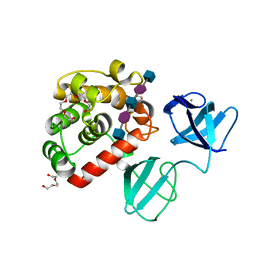 | |
7PJ4
 
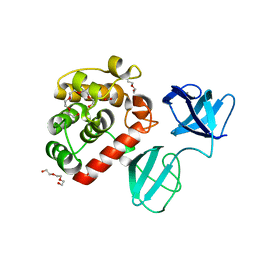 | |
7PL5
 
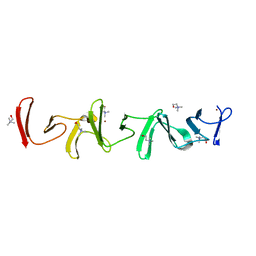 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
7POD
 
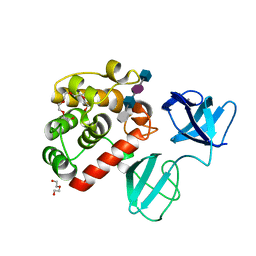 | |
7PJ3
 
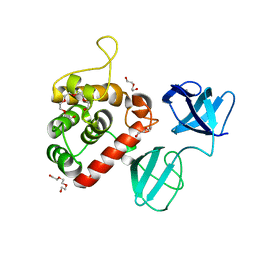 | |
8S5B
 
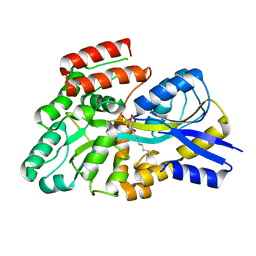 | | Crystal structure of the sulfoquinovosyl binding protein (SmoF) from A. tumefaciens sulfo-SMO pathway in complex with SQOctyl ligand | | Descriptor: | Sulfoquinovosyl glycerol-binding protein SmoF, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-octoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Sharma, M, Davies, G.J. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture-and-release of a sulfoquinovose-binding protein on sulfoquinovose-modified agarose.
Org.Biomol.Chem., 22, 2024
|
|
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
