7WI1
 
 | |
1YNX
 
 | |
7BZ4
 
 | |
7BYQ
 
 | |
7BZ1
 
 | |
7BZI
 
 | |
7BZ3
 
 | |
1XNH
 
 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | 分子名称: | NH(3)-dependent NAD(+) synthetase | | 著者 | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | 登録日 | 2004-10-05 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1Q3P
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | 分子名称: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | 著者 | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | 登録日 | 2003-07-31 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1XHK
 
 | | Crystal structure of M. jannaschii Lon proteolytic domain | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative protease La homolog, SULFATE ION | | 著者 | Im, Y.J, Na, Y, Kang, G.B, Rho, S.-H, Kim, M.-K, Lee, J.H, Chung, C.H, Eom, S.H. | | 登録日 | 2004-09-20 | | 公開日 | 2004-10-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases.
J.Biol.Chem., 279, 2004
|
|
1Q3O
 
 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | 分子名称: | BROMIDE ION, Shank1 | | 著者 | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | 登録日 | 2003-07-31 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1XNG
 
 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | 著者 | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | 登録日 | 2004-10-05 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
3TJ5
 
 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-N; residues 412-434) from Rickettsia rickettsii | | 分子名称: | Antigenic heat-stable 120 kDa protein, GLYCEROL, Vinculin | | 著者 | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | 登録日 | 2011-08-23 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3TJ6
 
 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-C; residues 812-835) from Rickettsia rickettsii | | 分子名称: | Antigenic heat-stable 120 kDa protein, Vinculin | | 著者 | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | 登録日 | 2011-08-23 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3SMZ
 
 | |
7YC0
 
 | | Acetylesterase (LgEstI) W.T. | | 分子名称: | ACETATE ION, Alpha/beta hydrolase, CHLORIDE ION | | 著者 | Do, H, Lee, J.H. | | 登録日 | 2022-06-30 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
7YC4
 
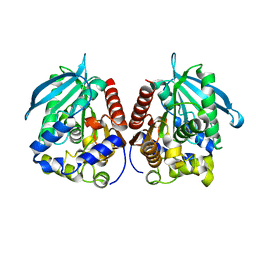 | | Acetylesterase (LgEstI) F207A | | 分子名称: | Alpha/beta hydrolase | | 著者 | Do, H, Lee, J.H. | | 登録日 | 2022-06-30 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
3NAH
 
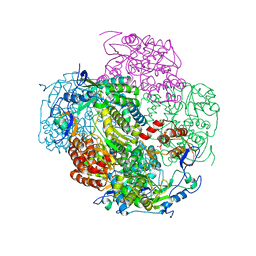 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | 分子名称: | RNA dependent RNA polymerase, SULFATE ION | | 著者 | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | 登録日 | 2010-06-02 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
3NAI
 
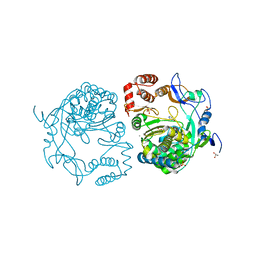 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | 分子名称: | 5-FLUOROURACIL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | 登録日 | 2010-06-02 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
7CS1
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Neomycin | | 分子名称: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, NEOMYCIN | | 著者 | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | 登録日 | 2020-08-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.966 Å) | | 主引用文献 | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSI
 
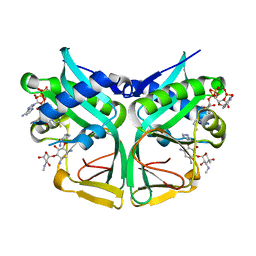 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Sisomicin | | 分子名称: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside 2'-N-acetyltransferase, COENZYME A | | 著者 | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | 登録日 | 2020-08-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CSJ
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Gentamicin | | 分子名称: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, gentamicin C1 | | 著者 | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | 登録日 | 2020-08-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.168 Å) | | 主引用文献 | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CRM
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-APO Structure | | 分子名称: | 1,2-ETHANEDIOL, Aminoglycoside 2'-N-acetyltransferase | | 著者 | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | 登録日 | 2020-08-13 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.487 Å) | | 主引用文献 | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7CS0
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Paromomycin | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Aminoglycoside 2'-N-acetyltransferase, ... | | 著者 | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | 登録日 | 2020-08-14 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
7DLS
 
 | | Cytochrome P450 (CYP105D18) complex with papaverine | | 分子名称: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Do, H, Lee, J.H. | | 登録日 | 2020-11-30 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Characterization of high-H 2 O 2 -tolerant bacterial cytochrome P450 CYP105D18: insights into papaverine N-oxidation.
Iucrj, 8, 2021
|
|
