4F15
 
 | | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Kim, K.H, Cho, K.J, Lee, J.H, Park, Y.H, Khan, T.G, Lee, J.Y, Kang, S.H, Alam, I. | | Deposit date: | 2012-05-06 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular basis of infectivity of 2009 pandemic H1N1 influenza A viruses
To be Published
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
8GJE
 
 | | HIV-1 Env subtype C CZA97.12 SOSIP.664 in complex with 3BNC117 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, J.H, Ward, A.B. | | Deposit date: | 2023-03-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan heterogeneity as a cause of the persistent fraction in HIV-1 neutralization.
Plos Pathog., 19, 2023
|
|
7C04
 
 | | Crystal structure of human Trap1 with DN203492 | | Descriptor: | 4-chloranyl-1-[[2-methoxy-4-(trifluoromethyl)phenyl]methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
7C05
 
 | | Crystal structure of human Trap1 with DN203495 | | Descriptor: | 1-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
7E8N
 
 | | Crystal structure of Type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554 | | Descriptor: | CITRIC ACID, Citrate synthase | | Authors: | Park, S.-H, Lee, C.W, Bae, D.-W, Lee, J.H. | | Deposit date: | 2021-03-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Int.J.Biol.Macromol., 183, 2021
|
|
5YX4
 
 | | Isoliquiritigenin-complexed Chalcone isomerase (S189A) from the Antarctic vascular plant Deschampsia Antarctica (DaCHI1) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Chalcone-flavonone isomerase family protein | | Authors: | Lee, C.W, Park, S, Lee, J.H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzymatic properties of chalcone isomerase from the Antarctic vascular plant Deschampsia antarctica Desv.
PLoS ONE, 13, 2018
|
|
5YX3
 
 | |
7BZ4
 
 | |
7BYQ
 
 | |
7BZ1
 
 | |
7BZI
 
 | |
7BZ3
 
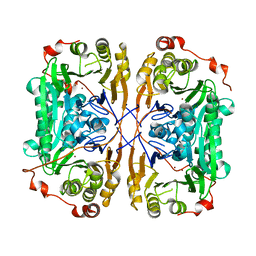 | |
3JTR
 
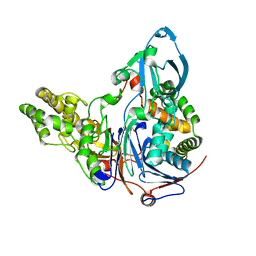 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3JTQ
 
 | | Mutations in Cephalosporin Acylase Affecting Stability and Autoproteolysis | | Descriptor: | GLYCEROL, Glutaryl 7-aminocephalosporanic acid acylase | | Authors: | Cho, K.J, Kim, J.K, Lee, J.H, Shin, H.J, Park, S.S, Kim, K.H. | | Deposit date: | 2009-09-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features of cephalosporin acylase reveal the basis of autocatalytic activation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
3L4F
 
 | | Crystal Structure of betaPIX Coiled-Coil Domain and Shank PDZ Complex | | Descriptor: | Rho guanine nucleotide exchange factor 7, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Im, Y.J, Kang, G.B, Lee, J.H, Song, H.E, Park, K.R, Kim, E, Song, W.K, Park, D, Eom, S.H. | | Deposit date: | 2009-12-19 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for asymmetric association of the betaPIX coiled coil and shank PDZ
J.Mol.Biol., 397, 2010
|
|
4NU3
 
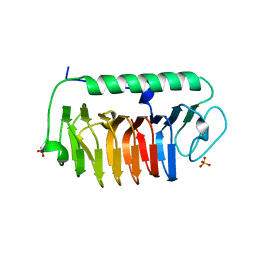 | | Crystal structure of mFfIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | SODIUM ION, SULFATE ION, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NU2
 
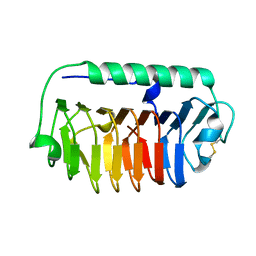 | | Crystal structure of an ice-binding protein (FfIBP) from the Antarctic bacterium, Flavobacterium frigoris PS1 | | Descriptor: | Antifreeze protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NUH
 
 | | Crystal structure of mLeIBP, a capping head region swapped mutant of ice-binding protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, ice-binding protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5D9V
 
 | |
5D9W
 
 | | Dehydroascorbate reductase (OsDHAR) complexed with ASA | | Descriptor: | ASCORBIC ACID, Dehydroascorbate reductase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2015-08-19 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6897 Å) | | Cite: | Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5D9T
 
 | |
7YC4
 
 | | Acetylesterase (LgEstI) F207A | | Descriptor: | Alpha/beta hydrolase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and biochemical analysis of acetylesterase (LgEstI) from Lactococcus garvieae.
Plos One, 18, 2023
|
|
