7OXR
 
 | |
7OXP
 
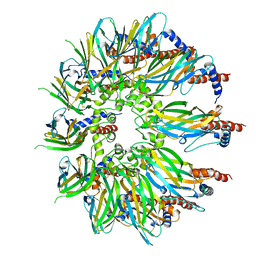 | | Cryo-EM structure of yeast Sei1 | | Descriptor: | BJ4_G0032880.mRNA.1.CDS.1,BJ4_G0032880.mRNA.1.CDS.1 | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex.
Nat Commun, 12, 2021
|
|
7P9V
 
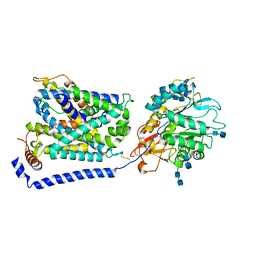 | | Cryo EM structure of System XC- | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
7P9U
 
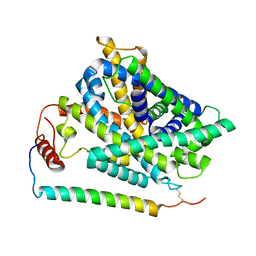 | | Cryo EM structure of System XC- in complex with glutamate | | Descriptor: | 4F2 cell-surface antigen heavy chain, Cystine/glutamate transporter, GLUTAMIC ACID | | Authors: | Parker, J.L, Deme, J.C, Lea, S.M, Newstead, S. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for redox control by the human cystine/glutamate antiporter system xc .
Nat Commun, 12, 2021
|
|
8SAH
 
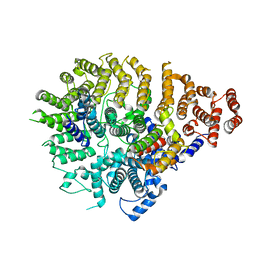 | | Huntingtin C-HEAT domain in complex with HAP40 | | Descriptor: | 40-kDa huntingtin-associated protein, Huntingtin | | Authors: | Harding, R.J, Deme, J.C, Alteen, M.G, Arrowsmith, C.H, Lea, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-31 | | Release date: | 2023-04-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Delineation of functional subdomains of Huntingtin protein and their interaction with HAP40.
Structure, 31, 2023
|
|
8SA2
 
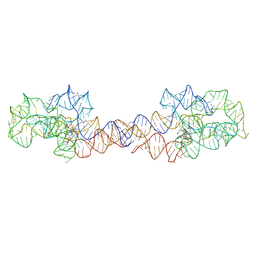 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
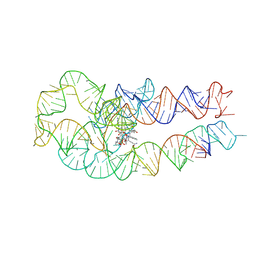 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
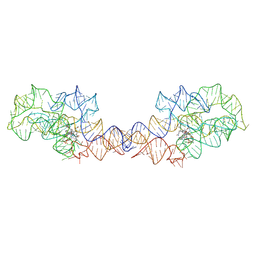 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
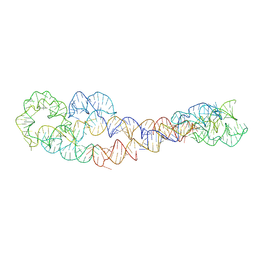 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
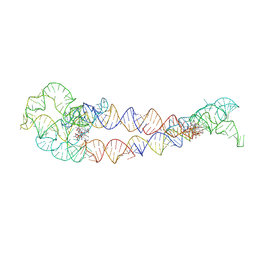 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
6H3I
 
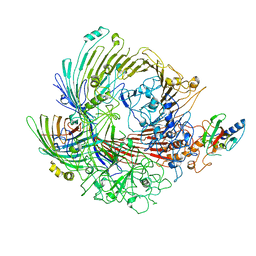 | | Structural snapshots of the Type 9 protein translocon | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, PorV, Protein involved in gliding motility SprA | | Authors: | Deme, J.C, Lea, S.M. | | Deposit date: | 2018-07-18 | | Release date: | 2018-11-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Type 9 secretion system structures reveal a new protein transport mechanism.
Nature, 564, 2018
|
|
6H3J
 
 | |
8GL6
 
 | |
8GLJ
 
 | |
8GLK
 
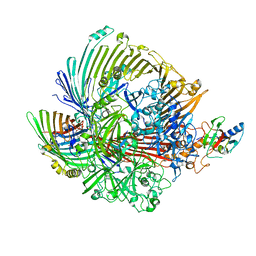 | |
8GLN
 
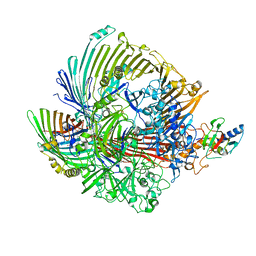 | |
8GLM
 
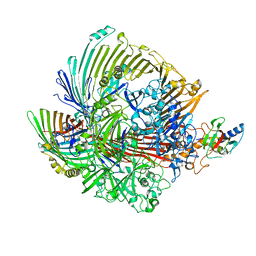 | |
8GL8
 
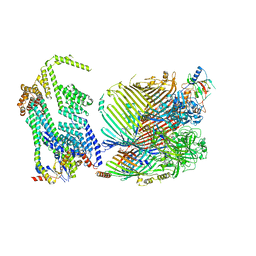 | |
8HC1
 
 | | CryoEM structure of Helicobacter pylori UreFD/urease complex | | Descriptor: | Urease accessory protein UreF, Urease accessory protein UreH, Urease subunit alpha, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
8HCN
 
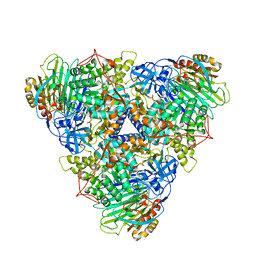 | | CryoEM Structure of Klebsiella pneumoniae UreD/urease complex | | Descriptor: | Urease accessory protein UreD, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Nim, Y.S, Fong, I.Y.H, Deme, J, Tsang, K.L, Caesar, J, Johnson, S, Wong, K.B, Lea, S.M. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Delivering a toxic metal to the active site of urease.
Sci Adv, 9, 2023
|
|
4TT9
 
 | |
7YXY
 
 | |
7YXX
 
 | |
7AKV
 
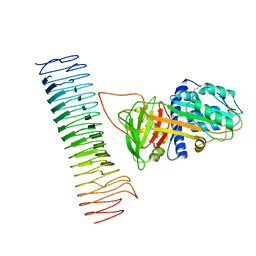 | | The cryo-EM structure of the Vag8-C1 inhibitor complex | | Descriptor: | Plasma protease C1 inhibitor, Vag8 | | Authors: | Johnson, S, Lea, S.M, Deme, J.C, Furlong, E, Dhillon, A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Mbio, 12, 2021
|
|
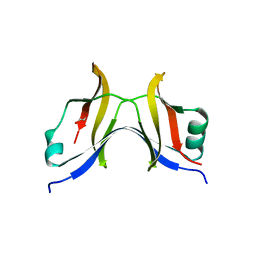
7ALJ
 
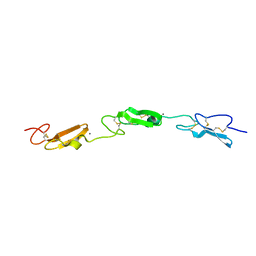 | | Structure of Drosophila Notch EGF domains 11-13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus Notch protein, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
