6P7H
 
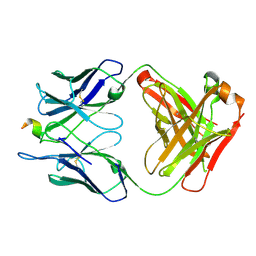 | |
3TGR
 
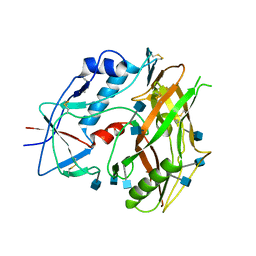 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TCL
 
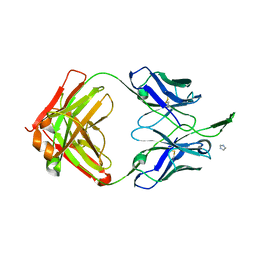 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6P60
 
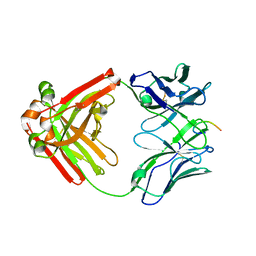 | |
6P8D
 
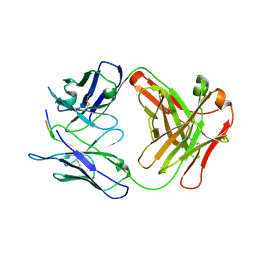 | |
3TGQ
 
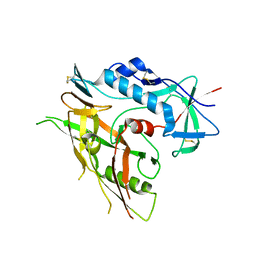 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6PEC
 
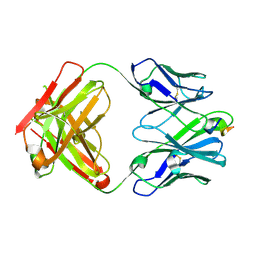 | |
3TGS
 
 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6PEF
 
 | |
3TIH
 
 | |
6PDS
 
 | | Vaccine-elicited NHP FP-targeting antibody 0PV-a.04 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, antibody 0PV-a.04 heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
3U36
 
 | | Crystal Structure of PG9 Fab | | Descriptor: | PG9 Fab heavy chain, PG9 Fab light chain, SULFATE ION | | Authors: | McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U4B
 
 | | CH04H/CH02L Fab P4 | | Descriptor: | CH02 Light chain, CH04 Heavy chain | | Authors: | Pancera, M, Louder, R, Mclellan, J.S, KWong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
6PDR
 
 | |
6PDU
 
 | | Vaccine-elicited NHP FP-targeting antibody 13N024-a.01 in complex with HIV fusion peptide (residue 512-519) | | Descriptor: | HIV-1 fusion peptide residue 512-519, antibody 13N024-a.01, heavy chain, ... | | Authors: | Xu, K, Liu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2019-06-19 | | Release date: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Modular recognition of antigens provides a mechanism that improves vaccine-elicited antibody-class frequencies
To Be Published
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
6Q0S
 
 | |
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3V7A
 
 | |
7LKB
 
 | |
7LKG
 
 | |
7LPN
 
 | |
7MFB
 
 | | Crystal structure of antibody 10E8v4 Fab - light chain H31F variant | | Descriptor: | Antibody 10E8v4 Fab heavy chain, Antibody 10E8v4 Fab light chain | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of HIV-1 Neutralizing Antibody 10E8 Delineate the Mechanistic Basis of Its Multi-Peak Behavior on Size-Exclusion Chromatography.
Antibodies, 10, 2021
|
|
