4DJJ
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution | | Descriptor: | PIMELIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution
To be Published
|
|
4DHW
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Adipic acid at 2.4 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase, hexanedioic acid | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Adipic acid at 2.4 Angstrom resolution
To be Published
|
|
4ERX
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with diethylene glycol at 2.5 Angstrom resolution
To be Published
|
|
8C4A
 
 | |
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
8IA6
 
 | |
8OH9
 
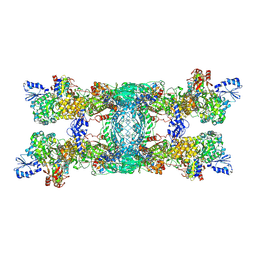 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 1) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Formate dehydrogenase-O, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
8OH5
 
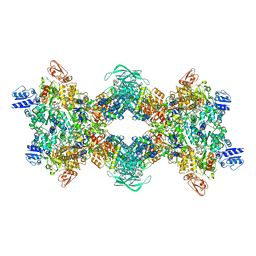 | | Cryo-EM structure of the electron bifurcating transhydrogenase StnABC complex from Sporomusa Ovata (state 2) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumar, A, Kremp, F, Mueller, V, Schuller, J.M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular architecture and electron transfer pathway of the Stn family transhydrogenase.
Nat Commun, 14, 2023
|
|
5ID2
 
 | |
1JYM
 
 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
4R60
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Ghosh, B, Are, V.N, Jamdar, S.N, Makde, R.D, Sharma, S.M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris
to be published
|
|
7RBH
 
 | | Human DNA polymerase beta crosslinked ternary complex 2 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBK
 
 | | Human DNA polymerase beta crosslinked complex, 40 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBL
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBJ
 
 | | Human DNA polymerase beta crosslinked complex, 30 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBI
 
 | | Human DNA polymerase beta crosslinked complex, 20 s Ca to Mg exchange | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBO
 
 | | Human DNA polymerase beta crosslinked complex, 60 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBM
 
 | | Human DNA polymerase beta crosslinked complex, 60 s Ca to Mn exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBN
 
 | | Human DNA polymerase beta crosslinked complex, 20 min Ca to Mg exchange | | Descriptor: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1CEH
 
 | | STRUCTURE AND FUNCTION OF THE CATALYTIC SITE MUTANT ASP99ASN OF PHOSPHOLIPASE A2: ABSENCE OF CONSERVED STRUCTURAL WATER | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Kumar, A, Sekharudu, C, Ramakrishnan, B, Dupureur, C.M, Zhu, H, Tsai, M.-D, Sundaralingam, M. | | Deposit date: | 1994-11-16 | | Release date: | 1995-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the catalytic site mutant Asp 99 Asn of phospholipase A2: absence of the conserved structural water.
Protein Sci., 3, 1994
|
|
3P2J
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Yadav, R, Sinha, M, Arora, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution
To be Published
|
|
7AG9
 
 | |
7C9B
 
 | | Crystal structure of dipeptidase-E from Xenopus laevis | | Descriptor: | Alpha-aspartyl dipeptidase, CALCIUM ION, SODIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
6JMI
 
 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
