5B35
 
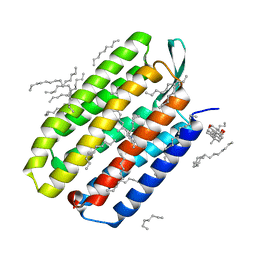 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B34
 
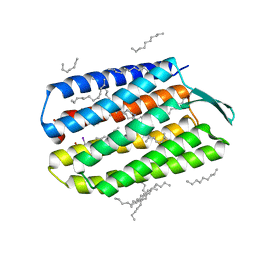 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXW
 
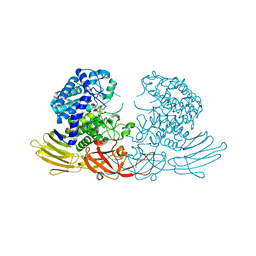 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | Descriptor: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7VDN
 
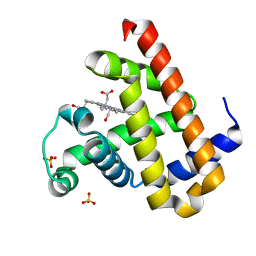 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7EXU
 
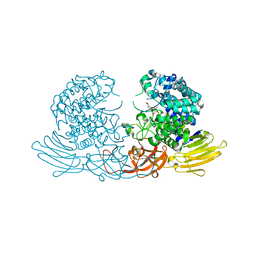 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | Descriptor: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
5GWA
 
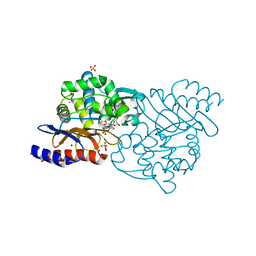 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5GS8
 
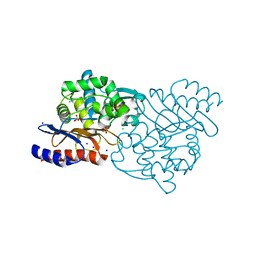 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2016-08-14 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
6JED
 
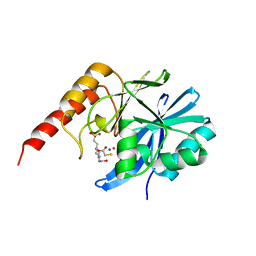 | | Crystal structure of IMP-1 metallo-beta-lactamase in a complex with MCR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Metallo-beta-lactamase type 2, SULFANYLACETIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
6K4T
 
 | | Crystal structure of SMB-1 metallo-beta-lactamase in a complex with TSA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-sulfanylbenzoic acid, Metallo-beta-lactamase, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
6K4X
 
 | | Crystal structure of SMB-1 metallo-beta-lactamase in a complex with ASB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-azanyl-2-sulfanyl-benzoic acid, Metallo-beta-lactamase, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KZL
 
 | | Crystal Structure Of NDM-1 Metallo-beta-lactamase In Complex With Inhibitor X2 | | Descriptor: | 2,5-diethyl-1-methyl-4-sulfamoyl-pyrrole-3-carboxylic acid, Beta-lactamase, SULFATE ION, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
6KXO
 
 | | Crystal Structure Of VIM-2 Metallo-beta-lactamase In Complex With Inhibitor NO9 | | Descriptor: | 2,5-dimethyl-4-sulfamoyl-furan-3-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
6KXI
 
 | |
6KZN
 
 | | Crystal Structure Of VIM-2 Metallo-beta-lactamase In Complex With Inhibitor X2 | | Descriptor: | 2,5-diethyl-1-methyl-4-sulfamoyl-pyrrole-3-carboxylic acid, Beta-lactamase class B VIM-2, FORMIC ACID, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
6LBL
 
 | | Crystal structure of IMP-1 metallo-beta-lactamase in complex with NO9 inhibitor | | Descriptor: | 2,5-dimethyl-4-sulfamoyl-furan-3-carboxylic acid, Metallo-beta-lactamase type 2, SODIUM ION, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-11-14 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Sulfamoyl Heteroarylcarboxylic Acids as Promising Metallo-beta-Lactamase Inhibitors for Controlling Bacterial Carbapenem Resistance.
Mbio, 11, 2020
|
|
2ZAE
 
 | | Crystal structure of protein Ph1601p in complex with protein Ph1771p of archaeal ribonuclease P from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, NITRATE ION, Ribonuclease P protein component 1, ... | | Authors: | Honda, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2007-10-04 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P.
J.Mol.Biol., 384, 2008
|
|
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
5XHF
 
 | | Crystal structure of Trastuzumab Fab fragment bearing p-azido-L-phenylalanine | | Descriptor: | polypeptide (H chain), polypeptide (L chain) | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
5XHG
 
 | | Crystal structure of Trastuzumab Fab fragment bearing Ne-(o-azidobenzyloxycarbonyl)-L-lysine | | Descriptor: | (2-azidophenyl)methyl hydrogen carbonate, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
1GVD
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GUU
 
 | | CRYSTAL STRUCTURE OF C-MYB R1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-01-30 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
5X5G
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
