5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
4YJ4
 
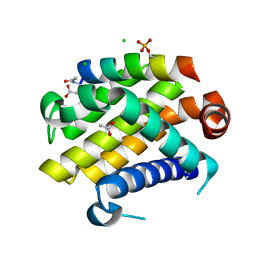 | | Crystal structure of Bcl-xL in complex with the BIM BH3 domain containing Ile155-to-Arg and Glu158-to-phosphoserine mutations | | Descriptor: | ACETATE ION, Bcl-2-like protein 1, BIM BH3 domain, ... | | Authors: | Ku, B, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-29 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of cell-survival activity of Akt into apoptotic death of cancer cells by two mutations on the BIM BH3 domain.
Cell Death Dis, 6, 2015
|
|
4X3N
 
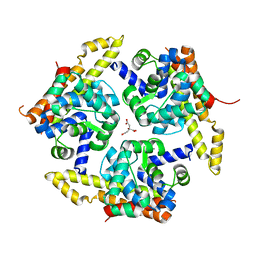 | | Crystal structure of 34 kDa F-actin bundling protein from Dictyostelium discoideum | | Descriptor: | CALCIUM ION, CITRIC ACID, Calcium-regulated actin-bundling protein | | Authors: | Kim, M.-K, Kim, J.-H, Kim, J.-S, Kang, S.-O. | | Deposit date: | 2014-12-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of the 34 kDa F-actin-bundling protein ABP34 from Dictyostelium discoideum.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1C7H
 
 | | CRYSTAL STRUCTURE OF A MUTANT R75A IN KETOSTEROID ISOMERASE FROM PSEDOMONAS PUTIDA BIOTYPE B | | Descriptor: | DELTA-5-3-KETOSTEROID ISOMERASE | | Authors: | Nam, G.H, Kim, D.H, Jang, D.S, Choi, G, Ha, N.C, Oh, B.H, Choi, K.Y. | | Deposit date: | 2000-02-19 | | Release date: | 2000-04-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Roles of active site aromatic residues in catalysis by ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 38, 1999
|
|
6XFP
 
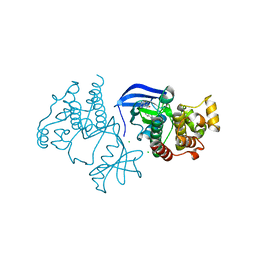 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
1DMQ
 
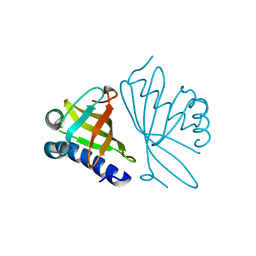 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
5AIR
 
 | | Structural analysis of mouse GSK3beta fused with LRP6 peptide. | | Descriptor: | Low-density lipoprotein receptor-related protein 6,Glycogen synthase kinase-3 beta, MALONATE ION | | Authors: | Kim, K.L. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Analysis of Mouse Gsk3 Beta Fused with Lrp6 Peptide
Biodesign, 3, 2015
|
|
8JWU
 
 | |
8JWS
 
 | | ePHD domain of PHD Finger Protein 7 (PHF7) | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 7, ZINC ION | | Authors: | Bang, I, Lee, H.S, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8JWJ
 
 | | PHD Finger Protein 7 (PHF7) in complex with UBE2D2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHD finger protein 7, ... | | Authors: | Lee, H.S, Bang, I, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
7CNC
 
 | | cystal structure of human ERH in complex with DGCR8 | | Descriptor: | Enhancer of rudimentary homolog, Microprocessor complex subunit DGCR8 | | Authors: | Li, F, Shen, S. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8.
Nucleic Acids Res., 48, 2020
|
|
7S10
 
 | | Crystal Structure of ascorbate peroxidase triple mutant: S160M, L203M, Q204M | | Descriptor: | L-ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poulos, T.L, Kim, J, Murarka, V. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.40000689 Å) | | Cite: | Computational analysis of the tryptophan cation radical energetics in peroxidase Compound I.
J.Biol.Inorg.Chem., 27, 2022
|
|
4TUG
 
 | | Crystal structure of MjMre11-DNA2 complex | | Descriptor: | DNA (5'-D(P*CP*TP*GP*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*CP*AP*GP*C)-3'), DNA double-strand break repair protein Mre11, ... | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
4TUI
 
 | | Crystal structure of MjMre11-DNA1 complex | | Descriptor: | DNA (5'-D(P*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*AP*G)-3'), DNA (5'-D(P*TP*GP*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*C)-3'), DNA double-strand break repair protein Mre11 | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
4TSV
 
 | |
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W01
 
 | |
1W02
 
 | |
5GWG
 
 | | Solution structure of rattusin | | Descriptor: | Defensin alpha-related sequence 1 | | Authors: | Lee, C.W, Min, H.J. | | Deposit date: | 2016-09-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges
Sci Rep, 7, 2017
|
|
1STZ
 
 | | Crystal structure of a hypothetical protein at 2.2 A resolution | | Descriptor: | Heat-inducible transcription repressor hrcA homolog | | Authors: | Liu, J, Adams, P.D, Shin, D.-H, Huang, C, Yokota, H, Jancarik, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a heat-inducible transcriptional repressor HrcA from Thermotoga maritima: structural insight into DNA binding and dimerization.
J.Mol.Biol., 350, 2005
|
|
6L6R
 
 | | Crystal structure of LRP6 E1E2-SOST complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Choi, H.-J, Kim, J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains.
Nat Commun, 11, 2020
|
|
6D3W
 
 | | Chromosomal trehalose-6-phosphate phosphatase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Hofmann, A, Cross, M, Park, S.-Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trehalose 6-phosphate phosphatases of Pseudomonas aeruginosa.
FASEB J., 32, 2018
|
|
6D3V
 
 | | Chromosomal trehalose-6-phosphate phosphatase from P. aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, NICKEL (II) ION, ... | | Authors: | Hofmann, A, Cross, M, Park, S.-Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trehalose 6-phosphate phosphatases of Pseudomonas aeruginosa.
FASEB J., 32, 2018
|
|
