4L39
 
 | | Crystal structure of GH3.12 from Arabidopsis thaliana in complex with AMPCPP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Zubieta, C, Jez, J.M, Brown, E, Marcellin, R, Kapp, U, Round, A, Westfall, C. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MAF
 
 | | Soybean ATP Sulfurylase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, ATP sulfurylase | | Authors: | Herrmann, J, Ravilious, G.E, McKinney, S.E, Westfall, C.S, Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and mechanism of soybean ATP sulfurylase and the committed step in plant sulfur assimilation.
J.Biol.Chem., 289, 2014
|
|
4N6A
 
 | | Soybean Serine Acetyltransferase Apoenzyme | | Descriptor: | PHOSPHATE ION, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4N6B
 
 | | Soybean Serine Acetyltransferase Complexed with CoA | | Descriptor: | COENZYME A, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4N69
 
 | | Soybean Serine Acetyltransferase Complexed with Serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4NJ6
 
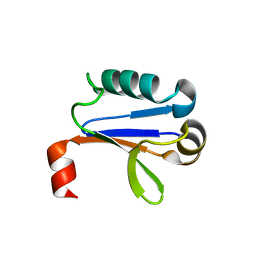 | | PB1 Domain of AtARF7 | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJ7
 
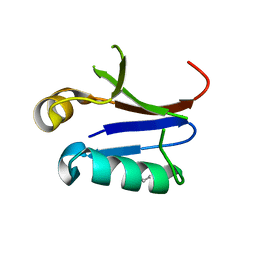 | | PB1 Domain of AtARF7 - SeMet Derivative | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OMZ
 
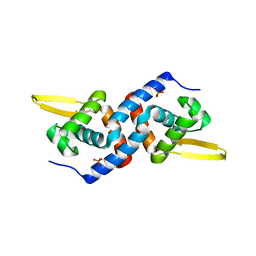 | |
4OMY
 
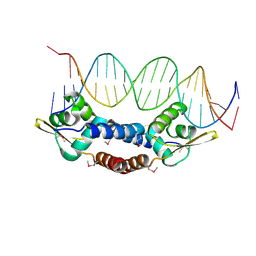 | |
4ON0
 
 | | Crystal Structure of NolR from Sinorhizobium fredii in complex with oligo AA DNA | | Descriptor: | DNA (5 -D(*TP*AP*AP*TP*CP*TP*CP*TP*TP*GP*GP*GP*AP*CP*TP*AP*CP*AP*AP*TP*TP*A)-3 ), DNA (5 -D(*TP*AP*TP*TP*AP*GP*AP*GP*AP*AP*CP*CP*CP*TP*GP*AP*TP*GP*TP*TP*AP*A)-3 ), NolR | | Authors: | Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for regulation of rhizobial nodulation and symbiosis gene expression by the regulatory protein NolR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PPU
 
 | |
4PPV
 
 | |
5T9F
 
 | | Prephenate Dehydrogenase N222D mutant from Soybean | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1, TYROSINE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2016-09-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
5T95
 
 | | Prephenate Dehydrogenase M219T, N222D mutant from Soybean | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1, TYROSINE | | Authors: | Holland, C.K, Jez, J.M. | | Deposit date: | 2016-09-09 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
5T9E
 
 | |
5CB6
 
 | | Structure of adenosine-5'-phosphosulfate kinase | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Herrmann, J, Jez, J.M. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Recapitulating the Structural Evolution of Redox Regulation in Adenosine 5'-Phosphosulfate Kinase from Cyanobacteria to Plants.
J.Biol.Chem., 290, 2015
|
|
5CB8
 
 | |
5TVS
 
 | | JMJD2A in complex with Ni(II) | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
5TVR
 
 | | JMJD2A in complex with Ni(II) and alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
3TSY
 
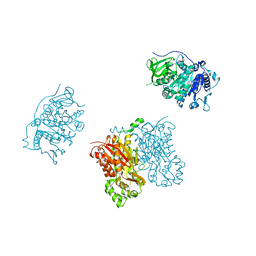 | | 4-Coumaroyl-CoA Ligase::Stilbene Synthase fusion protein | | Descriptor: | Fusion Protein 4-coumarate--CoA ligase 1, Resveratrol synthase | | Authors: | Yi, H, Jez, J.M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Kinetic Analysis of the Unnatural Fusion Protein 4-Coumaroyl-CoA Ligase::Stilbene Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
3UIE
 
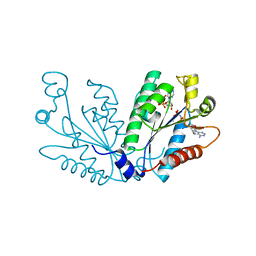 | |
3UJA
 
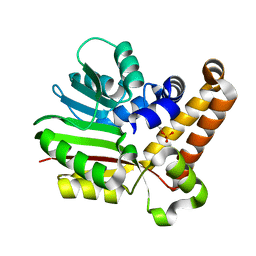 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.466 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJB
 
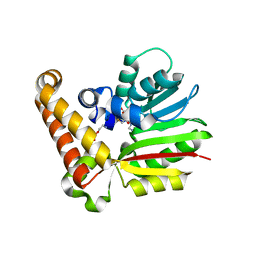 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAH and phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphoethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJ6
 
 | | SeMet Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAM and PO4 | | Descriptor: | PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJD
 
 | | Phosphoethanolamine methyltransferase mutant (Y19F) from Plasmodium falciparum in complex with phosphocholine | | Descriptor: | PHOSPHOCHOLINE, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
