1ZHZ
 
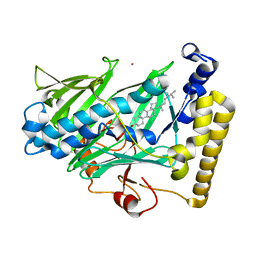 | | Structure of yeast oxysterol binding protein Osh4 in complex with ergosterol | | 分子名称: | ERGOSTEROL, KES1 protein, LEAD (II) ION | | 著者 | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | 登録日 | 2005-04-26 | | 公開日 | 2005-09-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHY
 
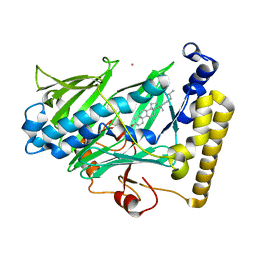 | | Structure of yeast oxysterol binding protein Osh4 in complex with cholesterol | | 分子名称: | CHOLESTEROL, KES1 protein, LEAD (II) ION | | 著者 | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | 登録日 | 2005-04-26 | | 公開日 | 2005-09-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1ZHT
 
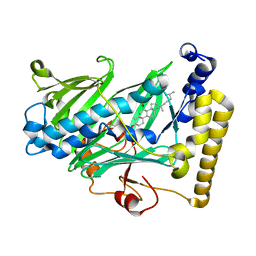 | | Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol | | 分子名称: | 7-HYDROXYCHOLESTEROL, KES1 protein | | 著者 | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | 登録日 | 2005-04-26 | | 公開日 | 2005-09-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
1Z2O
 
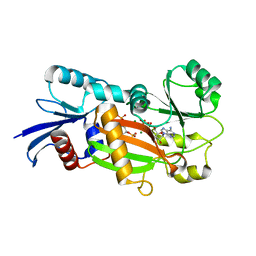 | | Inositol 1,3,4-trisphosphate 5/6-Kinase in complex with mg2+/ADP/Ins(1,3,4,6)P4 | | 分子名称: | (1S,3R,4R,6S)-1,3,4,6-TETRAPKISPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | 登録日 | 2005-03-08 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
1ZB1
 
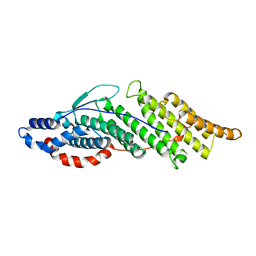 | | Structure basis for endosomal targeting by the Bro1 domain | | 分子名称: | BRO1 protein | | 著者 | Kim, J, Sitaraman, S, Hierro, A, Beach, B.M, Odorizzi, G, Hurley, J.H. | | 登録日 | 2005-04-07 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for endosomal targeting by the Bro1 domain.
Dev.Cell, 8, 2005
|
|
1JUQ
 
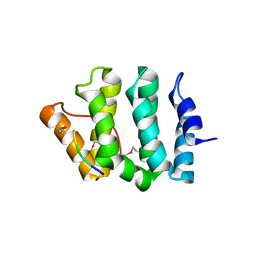 | | GGA3 VHS domain complexed with C-terminal peptide from cation-dependent Mannose 6-phosphate receptor | | 分子名称: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, Cation-dependent mannose-6-phosphate receptor | | 著者 | Misra, S, Puertollano, R, Bonifacino, J.S, Hurley, J.H. | | 登録日 | 2001-08-26 | | 公開日 | 2002-02-27 | | 最終更新日 | 2016-10-12 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains.
Nature, 415, 2002
|
|
1JPL
 
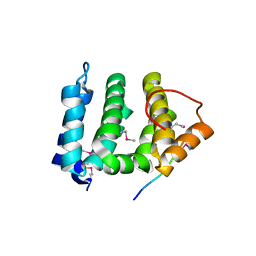 | | GGA3 VHS domain complexed with C-terminal peptide from cation-independent mannose 6-phosphate receptor | | 分子名称: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, Cation-Independent Mannose 6-phosphate receptor | | 著者 | Misra, S, Puertollano, R, Bonifacino, J.S, Hurley, J.H. | | 登録日 | 2001-08-02 | | 公開日 | 2002-02-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains.
Nature, 415, 2002
|
|
1LF8
 
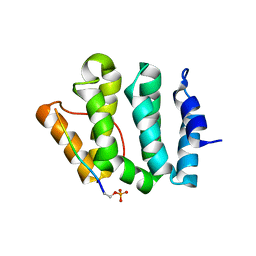 | | Complex of GGA3-VHS Domain and CI-MPR C-terminal Phosphopeptide | | 分子名称: | ADP-ribosylation factor binding protein GGA3, Cation-independent mannose-6-phosphate receptor | | 著者 | Kato, Y, Misra, S, Puertollano, R, Hurley, J.H, Bonifacino, J.S. | | 登録日 | 2002-04-10 | | 公開日 | 2002-06-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism.
Nat.Struct.Biol., 9, 2002
|
|
1MN3
 
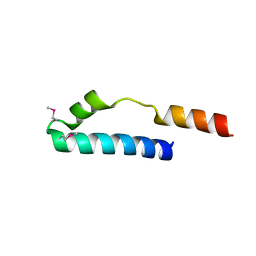 | | Cue domain of yeast Vps9p | | 分子名称: | Vacuolar protein sorting-associated protein VPS9 | | 著者 | Prag, G, Misra, S, Jones, E, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | 登録日 | 2002-09-04 | | 公開日 | 2003-06-10 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p
Cell(Cambridge,Mass.), 113, 2003
|
|
1MLV
 
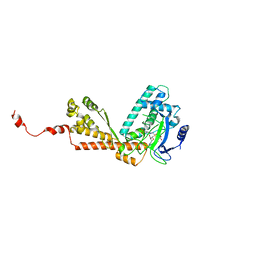 | | Structure and Catalytic Mechanism of a SET Domain Protein Methyltransferase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ribulose-1,5 biphosphate carboxylase/oxygenase large subunit N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Trievel, R.C, Beach, B.M, Dirk, L.M.A, Houtz, R.L, Hurley, J.H. | | 登録日 | 2002-08-30 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and catalytic mechanism of a SET domain protein methyltransferase.
Cell(Cambridge,Mass.), 111, 2002
|
|
1NWM
 
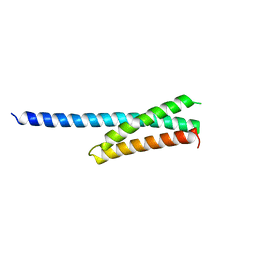 | | GAT domain of human GGA1 | | 分子名称: | ADP-ribosylation factor binding protein GGA1 | | 著者 | Suer, S, Misra, S, Saidi, L.F, Hurley, J.H. | | 登録日 | 2003-02-06 | | 公開日 | 2003-03-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the GAT domain of human GGA1: a syntaxin amino-terminal domain fold in an endosomal trafficking adaptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P0Y
 
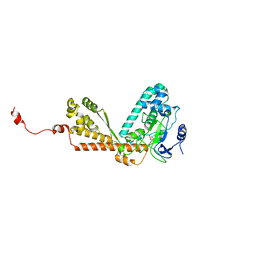 | | Crystal structure of the SET domain of LSMT bound to MeLysine and AdoHcy | | 分子名称: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | 著者 | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | 登録日 | 2003-04-11 | | 公開日 | 2003-07-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1P4U
 
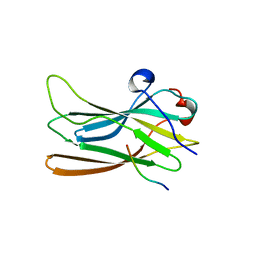 | | CRYSTAL STRUCTURE OF GGA3 GAE DOMAIN IN COMPLEX WITH RABAPTIN-5 PEPTIDE | | 分子名称: | ADP-ribosylation factor binding protein GGA3, Rabaptin-5 | | 著者 | Miller, G.J, Mattera, R, Bonifacino, J.S, Hurley, J.H. | | 登録日 | 2003-04-24 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | RECOGNITION OF ACCESSORY PROTEIN MOTIFS BY THE GAMMA-ADAPTIN EAR DOMAIN OF GGA3
Nat.Struct.Biol., 10, 2003
|
|
7JTL
 
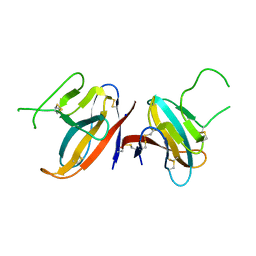 | | Structure of SARS-CoV-2 ORF8 accessory protein | | 分子名称: | ORF8 protein, SODIUM ION | | 著者 | Flower, T.G, Buffalo, C.Z, Hooy, R.M, Allaire, M, Ren, X, Hurley, J.H. | | 登録日 | 2020-08-18 | | 公開日 | 2020-08-26 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5PTP
 
 | |
5L1Z
 
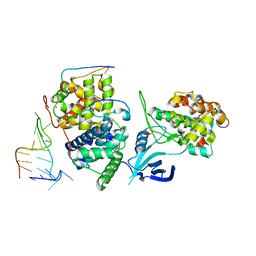 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | 分子名称: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | 著者 | Schulze-Gahmen, U, Hurley, J. | | 登録日 | 2016-07-29 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (5.9 Å) | | 主引用文献 | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
7R4H
 
 | |
7UX2
 
 | |
7UXC
 
 | |
7UXH
 
 | |
4N7H
 
 | | Crystal Structure of the Complex of 3rd WW domain of Human Nedd4 and 1st PPXY Motif of ARRDC3 | | 分子名称: | Arrestin domain-containing protein 3, E3 ubiquitin-protein ligase NEDD4, GUANIDINE | | 著者 | Qi, S, O'Hayre, M, Gutkind, J.S, Hurley, J. | | 登録日 | 2013-10-15 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3).
J.Biol.Chem., 289, 2014
|
|
4N7F
 
 | |
1BL5
 
 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | 分子名称: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | 著者 | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | 登録日 | 1998-07-23 | | 公開日 | 1999-05-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
4R7X
 
 | |
1IDE
 
 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | 分子名称: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | 著者 | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | 登録日 | 1995-01-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
