7A4F
 
 | |
7A4G
 
 | |
7A4I
 
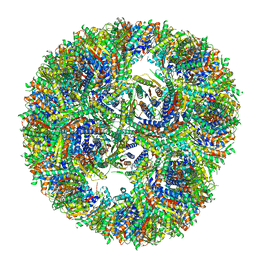
 | | Aquifex aeolicus lumazine synthase-derived nucleocapsid variant NC-3 | | Descriptor: | Antitermination protein N,6,7-dimethyl-8-ribityllumazine synthase,6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Tetter, S, Hilvert, D. | | Deposit date: | 2020-08-19 | | Release date: | 2021-06-02 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (7.04 Å) | | Cite: | Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.
Science, 372, 2021
|
|
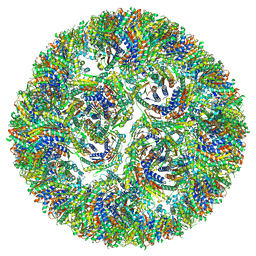
7A4H
 
 | |
6ZV9
 
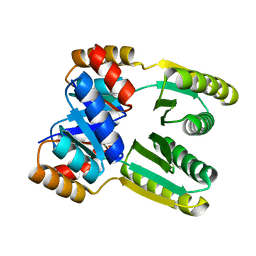 | | Terbium(III)-bound de novo TIM barrel-ferredoxin fold fusion dimer with 4-glutamate binding site and tryptophan antenna (TFD-EE N6W) | | Descriptor: | 1,2-ETHANEDIOL, TERBIUM(III) ION, TFD-EE | | Authors: | Caldwell, S, Haydon, I, Piperidou, N, Huang, P, Hilvert, D, Baker, D, Zeymer, C. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
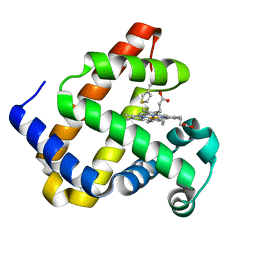
6Z4R
 
 | |
6Z4T
 
 | |
7TXV
 
 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 E82Q with ATP and 16x(Asp-Arg) | | Descriptor: | 16x(Asp-Arg), ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Nat Commun, 13, 2022
|
|
7TXU
 
 | | Cyanophycin synthetase 1 from Synechocystis sp. UTEX2470 with ATP and 16x(Asp-Arg) | | Descriptor: | 16x(Asp-Arg), ADENOSINE-5'-TRIPHOSPHATE, Cyanophycin synthase, ... | | Authors: | Sharon, I, Grogg, M, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A cryptic third active site in cyanophycin synthetase creates primers for polymerization
Nat Commun, 13, 2022
|
|
8F7I
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P43212 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
8F7F
 
 | | The condensation domain of surfactin A synthetase C in space group P43212 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
8F7H
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P212121 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
8F7G
 
 | | The condensation domain of surfactin A synthetase C in space group P212121 | | Descriptor: | GLYCEROL, Surfactin synthetase | | Authors: | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 2024
|
|
4PA8
 
 | | Crystal structure of a de novo retro-aldolase catalyzing asymmetric Michael additions, with a covalently bound product analog | | Descriptor: | (3R)-3-(4-methoxyphenyl)-5-oxohexanenitrile, GLYCEROL, SULFATE ION, ... | | Authors: | Beck, T, Garrabou Pi, X, Hilvert, D. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Promiscuous De Novo Retro-Aldolase Catalyzes Asymmetric Michael Additions via Schiff Base Intermediates.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5L86
 
 | | engineered ascorbate peroxidise | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hayashi, T, Mittl, P, Hilvert, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Chemically Programmed Proximal Ligand Enhances the Catalytic Properties of a Heme Enzyme.
J. Am. Chem. Soc., 138, 2016
|
|
3ZP7
 
 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with chorismate and prephenate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, CHORISMATE MUTASE AROH, PREPHENIC ACID | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZP4
 
 | | Arg90Cit chorismate mutase of Bacillus subtilis in complex with a transition state analog | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-26 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZOP
 
 | | Arg90Cit chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
 | | Structure of the engineered retro-aldolase RA95.5-5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
6F17
 
 | | Structure of Mb NMH H64V, V68A mutant resting state | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
6F19
 
 | | Structure of Mb NMH H64V, V68A mutant complex with EDA incubated at room temperature for 5 min | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
