7CSW
 
 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
5U8F
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U69
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U2M
 
 | | Crystal structure of human NAMPT with A-1293201 | | Descriptor: | N-[4-({[(3S)-oxolan-3-yl]methyl}carbamoyl)phenyl]-1,3-dihydro-2H-isoindole-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
5U8A
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U6D
 
 | |
5U2N
 
 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
4HVI
 
 | |
4HVH
 
 | |
4HVG
 
 | |
7VSQ
 
 | |
7VSP
 
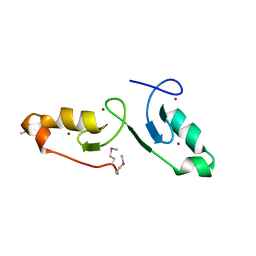 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
4HVD
 
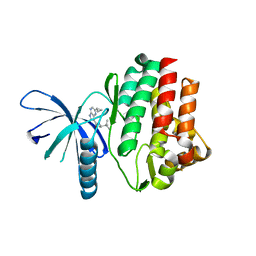 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
6NWS
 
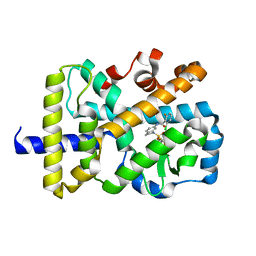 | | RORgamma Ligand Binding Domain | | Descriptor: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWU
 
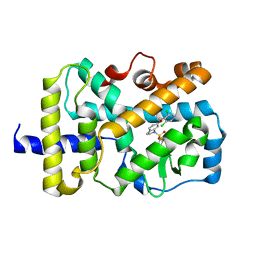 | | RORgamma Ligand Binding Domain | | Descriptor: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
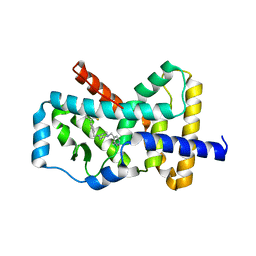 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6Z4Q
 
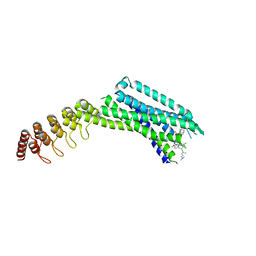 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule inverse agonist SR142948A | | Descriptor: | 2-[[5-(2,6-dimethoxyphenyl)-1-[4-[3-(dimethylamino)propyl-methyl-carbamoyl]-2-propan-2-yl-phenyl]pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZIN
 
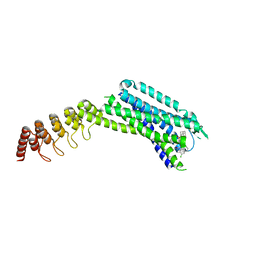 | | Crystal structure of the neurotensin receptor 1 in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZA8
 
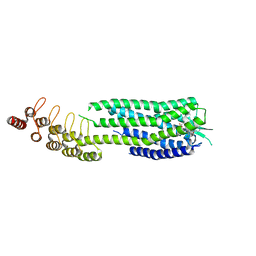 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4V
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with NTS8-13 | | Descriptor: | ARG-PRO-TYR-ILE-LEU, Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6YVR
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the peptide full agonist NTS8-13 | | Descriptor: | Neurotensin receptor type 1,Neurotensin receptor type 1,DARPin crystallisation chaperone, neurotensin NTS8-13 (full agonist), nonyl beta-D-glucopyranoside | | Authors: | Deluigi, M, Merklinger, L, Hilge, M, Ernst, P, Klipp, A, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-04-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z4S
 
 | | Crystal structure of the neurotensin receptor 1 (NTSR1-H4bmx) in complex with the small molecule inverse agonist SR48692 | | Descriptor: | 2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]adamantane-2-carboxylic acid, Neurotensin receptor type 1,DARPin,HRV 3C protease recognition sequence | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-05-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
