3CRO
 
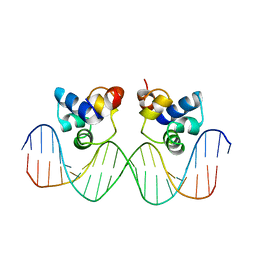 | |
6U1X
 
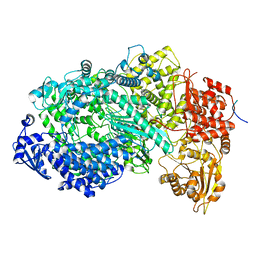 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | 分子名称: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | 登録日 | 2019-08-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
6UEB
 
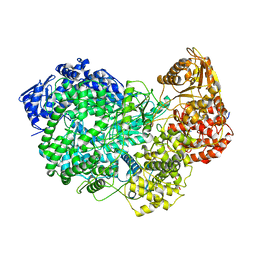 | |
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Chen, B, Harrison, S.C. | | 登録日 | 2019-10-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6UPH
 
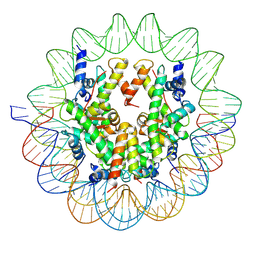 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | 分子名称: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | 著者 | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | 登録日 | 2019-10-17 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
6V05
 
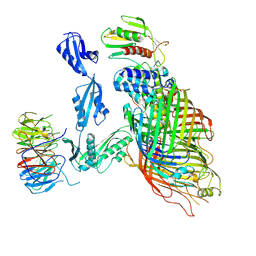 | | Cryo-EM structure of a substrate-engaged Bam complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | 著者 | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
1SID
 
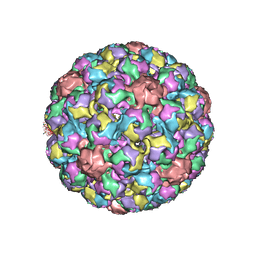 | | MURINE POLYOMAVIRUS COMPLEXED WITH 3'SIALYL LACTOSE | | 分子名称: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, POLYOMAVIRUS COAT PROTEIN VP1 | | 著者 | Stehle, T, Harrison, S.C. | | 登録日 | 1995-12-12 | | 公開日 | 1996-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments.
Structure, 4, 1996
|
|
7N64
 
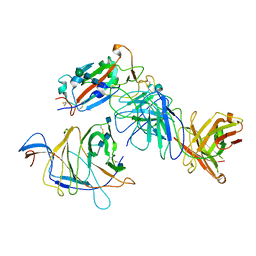 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | 著者 | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | 登録日 | 2021-06-07 | | 公開日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
7N62
 
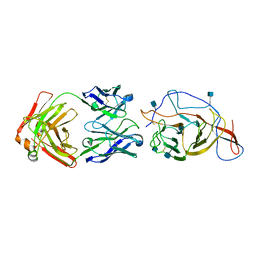 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | 著者 | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | 登録日 | 2021-06-07 | | 公開日 | 2021-08-04 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
2I3S
 
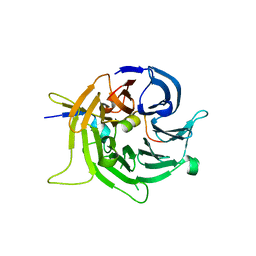 | | Bub3 complex with Bub1 GLEBS motif | | 分子名称: | Cell cycle arrest protein, Checkpoint serine/threonine-protein kinase | | 著者 | Larsen, N.A, Harrison, S.C. | | 登録日 | 2006-08-20 | | 公開日 | 2007-01-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2I3T
 
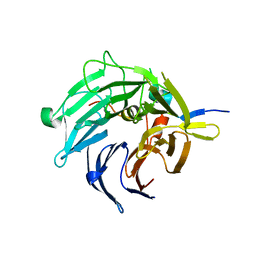 | |
8V10
 
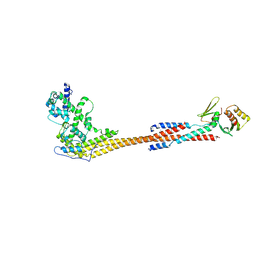 | |
8V11
 
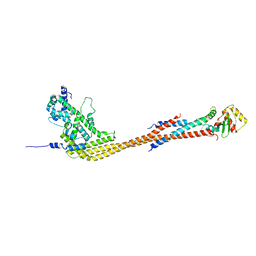 | |
2YFW
 
 | |
2YFV
 
 | |
5A22
 
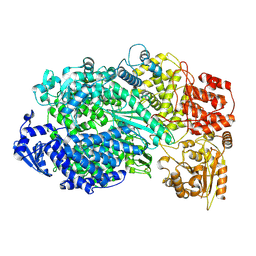 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | 分子名称: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | 著者 | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | 登録日 | 2015-05-06 | | 公開日 | 2015-08-19 | | 最終更新日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
2B6O
 
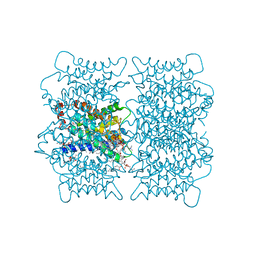 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | 著者 | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | 登録日 | 2005-10-03 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | 主引用文献 | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
6CBJ
 
 | |
1PER
 
 | |
6CFZ
 
 | |
6P7W
 
 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6CBP
 
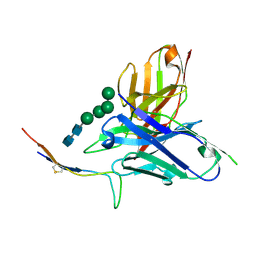 | |
6P7V
 
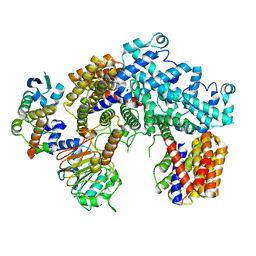 | | Structure of the K. lactis CBF3 core | | 分子名称: | Cep3, Ctf13, Skp1 | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
6PP7
 
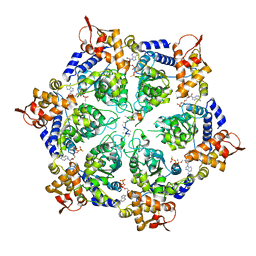 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6P7X
 
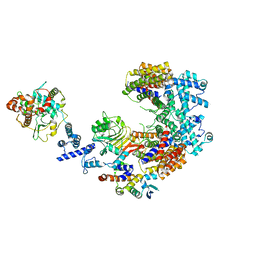 | | Structure of the K. lactis CBF3 core - Ndc10 D1D2 complex | | 分子名称: | Cep3, Ctf13, Ndc10, ... | | 著者 | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | 登録日 | 2019-06-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
