7F8X
 
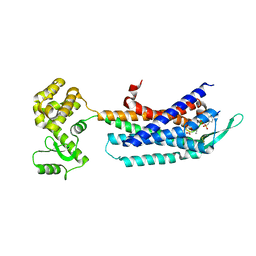 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7EE2
 
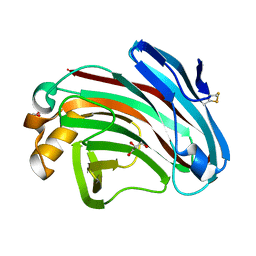 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | Descriptor: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37011635 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEE
 
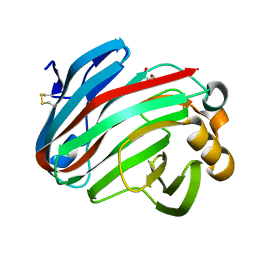 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | Descriptor: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.660792 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEJ
 
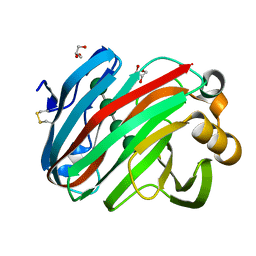 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | Descriptor: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2021-03-18 | | Release date: | 2022-03-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47798049 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
2FBX
 
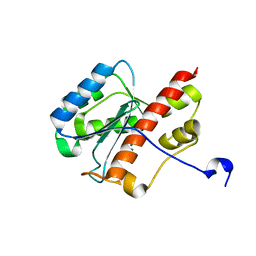 | | WRN exonuclease, Mg complex | | Descriptor: | MAGNESIUM ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FC0
 
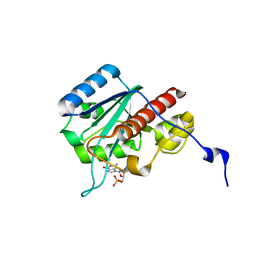 | | WRN exonuclease, Mn dGMP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J, Tainer, J.A. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBY
 
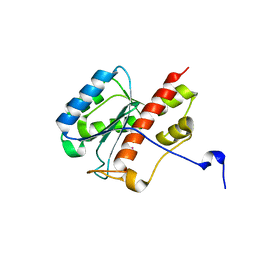 | | WRN exonuclease, Eu complex | | Descriptor: | EUROPIUM (III) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBV
 
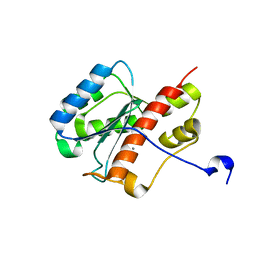 | | WRN exonuclease, Mn complex | | Descriptor: | MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBT
 
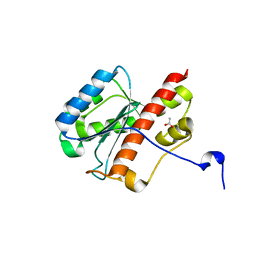 | | WRN exonuclease | | Descriptor: | ACETIC ACID, Werner syndrome helicase | | Authors: | Perry, J.J. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4TQ1
 
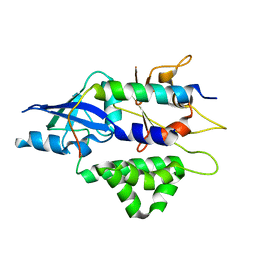 | | Crystal structure of human ATG5-TECAIR | | Descriptor: | Autophagy protein 5, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
4TQ0
 
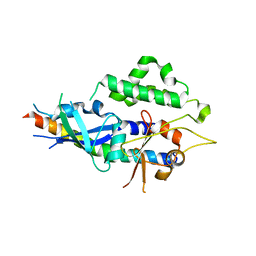 | | Crystal structure of human ATG5-ATG16N69 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
6L3A
 
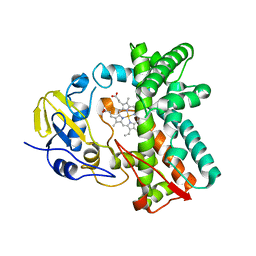 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6L39
 
 | | Cytochrome P450 107G1 (RapN) | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
8IHM
 
 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHT
 
 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
7XMR
 
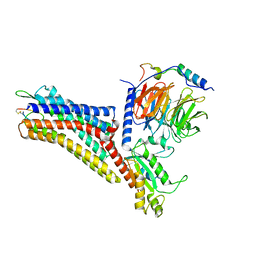 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
5YGI
 
 | |
5YVK
 
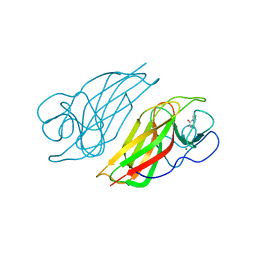 | | Crystal structure of a cyclase Famc1 from Fischerella ambigua UTEX 1903 | | Descriptor: | CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z54
 
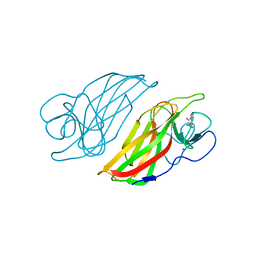 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-17 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YVP
 
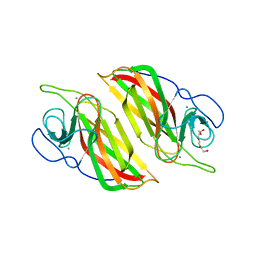 | | Crystal structure of an apo form cyclase Filc1 from Fischerella sp. TAU | | Descriptor: | CALCIUM ION, TETRAETHYLENE GLYCOL, cyclase A | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z53
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with cyclo-L-Arg-D-Pro | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, SULFATE ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-16 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YVL
 
 | | Crystal structure of a cyclase Hpiu5 from Fischerella sp. ATCC 43239 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, cyclase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-11-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
