2REW
 
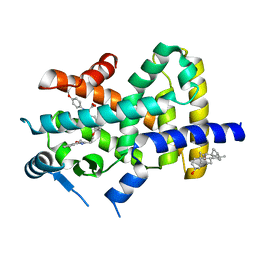 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | Descriptor: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | Authors: | Muckelbauer, J. | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
5F4H
 
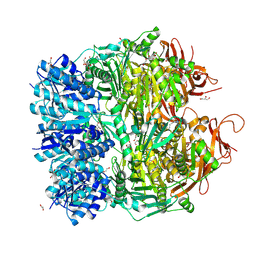 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, DuPrez, K.T, Doukov, T.I, Shen, Y, Fan, L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
J. Mol. Biol., 429, 2017
|
|
7TE8
 
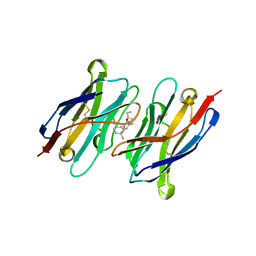 | | CA14-CBD-DB21 ternary complex | | Descriptor: | CA14, DB21, cannabidiol | | Authors: | Cao, S, Zheng, N. | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Defining molecular glues with a dual-nanobody cannabidiol sensor.
Nat Commun, 13, 2022
|
|
4M5F
 
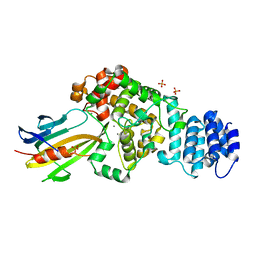 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5ZO4
 
 | | inactive state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5YWW
 
 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, Yuan, Z, Han, X, DuPrez, K, Shen, Y, Fan, L. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The archaeal ATPase PINA interacts with the helicase Hjm via its carboxyl terminal KH domain remodeling and processing replication fork and Holliday junction.
Nucleic Acids Res., 46, 2018
|
|
5ZO3
 
 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO5
 
 | | active state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
6A06
 
 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
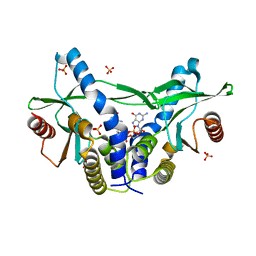 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A05
 
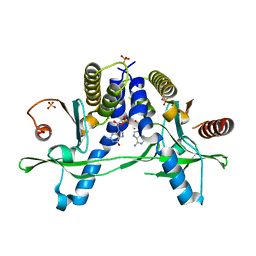 | | Structure of pSTING complex | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
3SP4
 
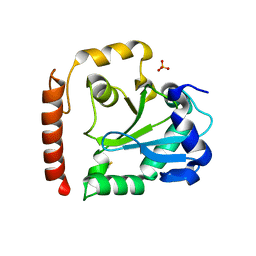 | | Crystal structure of aprataxin ortholog Hnt3 from Schizosaccharomyces pombe | | Descriptor: | Aprataxin-like protein, SULFATE ION, ZINC ION | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SPL
 
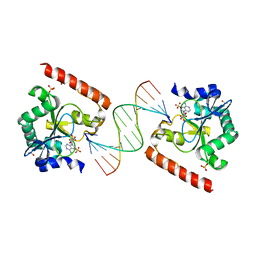 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), ... | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SPD
 
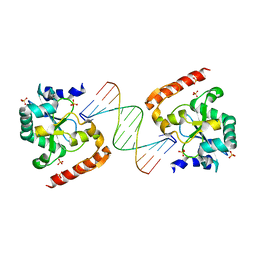 | | Crystal structure of aprataxin ortholog Hnt3 in complex with DNA | | Descriptor: | Aprataxin-like protein, DNA (5'-D(*GP*TP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*GP*AP*G)-3'), DNA (5'-D(*TP*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Gong, Y, Zhu, D, Ding, J, Dou, C, Ren, X, Jiang, T, Wang, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5'-adenylated DNA
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NNJ
 
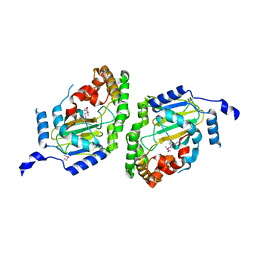 | | Halogenase domain from CurA module (apo Hal) | | Descriptor: | CurA | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNL
 
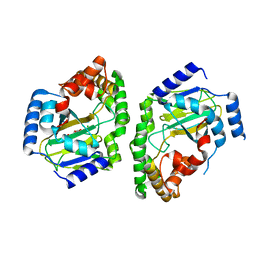 | | Halogenase domain from CurA module (crystal form III) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNF
 
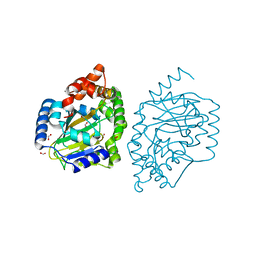 | | Halogenase domain from CurA module with Fe, chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNM
 
 | |
3DD4
 
 | |
3DOR
 
 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DJA
 
 | | Crystal Structure of cpaf solved with MAD | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-06-22 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPN
 
 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
