3MAU
 
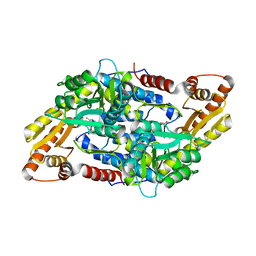 | | Crystal structure of StSPL in complex with phosphoethanolamine | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, PHOSPHATE ION, sphingosine-1-phosphate lyase, ... | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-24 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MAD
 
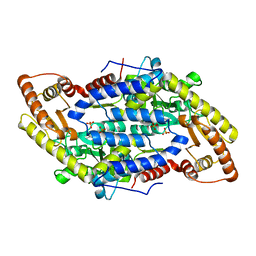 | |
3MBB
 
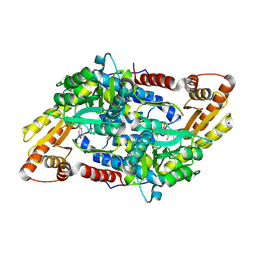 | | Crystal structure of StSPL - apo form, after treatment with semicarbazide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ... | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-25 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MC6
 
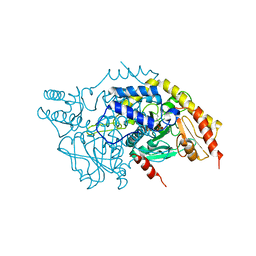 | | Crystal structure of ScDPL1 | | Descriptor: | PHOSPHATE ION, Sphingosine-1-phosphate lyase | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
3MAF
 
 | | Crystal structure of StSPL (asymmetric form) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, sphingosine-1-phosphate lyase | | Authors: | Bourquin, F, Grutter, M.G, Capitani, G. | | Deposit date: | 2010-03-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Structure and Function of Sphingosine-1-Phosphate Lyase, a Key Enzyme of Sphingolipid Metabolism.
Structure, 18, 2010
|
|
4DB6
 
 | | Designed Armadillo repeat protein (YIIIM3AII) | | Descriptor: | Armadillo repeat protein | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB8
 
 | | Designed Armadillo-repeat Protein | | Descriptor: | Armadillo-repeat Protein, MAGNESIUM ION | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DBA
 
 | | Designed Armadillo repeat protein (YIIM3AII) | | Descriptor: | Designed Armadillo repeat protein, YIIM3AII, GLYCEROL | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4DB9
 
 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
1PMM
 
 | | Crystal structure of Escherichia coli GadB (low pH) | | Descriptor: | ACETIC ACID, Glutamate decarboxylase beta, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
1PYO
 
 | |
1PMO
 
 | | Crystal structure of Escherichia coli GadB (neutral pH) | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutamate decarboxylase beta | | Authors: | Capitani, G, De Biase, D, Aurizi, C, Gut, H, Bossa, F, Grutter, M.G. | | Deposit date: | 2003-06-11 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of escherichia coli glutamate
decarboxylase
Embo J., 22, 2003
|
|
2BKG
 
 | | Crystal structure of E3_19 a designed ankyrin repeat protein | | Descriptor: | SYNTHETIC CONSTRUCT ANKYRIN REPEAT PROTEIN E3_19 | | Authors: | Binz, H.K, Kohl, A, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2005-02-16 | | Release date: | 2006-06-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Consensus-Designed Ankyrin Repeat Protein: Implications for Stability
Proteins: Struct., Funct., Bioinf., 65, 2006
|
|
1PN5
 
 | | NMR structure of the NALP1 Pyrin domain (PYD) | | Descriptor: | NACHT-, LRR- and PYD-containing protein 2 | | Authors: | Hiller, S, Kohl, A, Fiorito, F, Herrmann, T, Wider, G, Tschopp, J, Grutter, M.G, Wuthrich, K. | | Deposit date: | 2003-06-12 | | Release date: | 2003-10-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain
Structure, 11, 2003
|
|
2BKK
 
 | | Crystal structure of Aminoglycoside Phosphotransferase APH(3')-IIIa in complex with the inhibitor AR_3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE, DESIGNED ANKYRIN REPEAT INHIBITOR AR_3A, ... | | Authors: | Kohl, A, Amstutz, P, Parizek, P, Binz, H.K, Briand, C, Capitani, G, Forrer, P, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allosteric Inhibition of Aminoglycoside Phosphotransferase by a Designed Ankyrin Repeat Protein
Structure, 13, 2005
|
|
2CJX
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-09 | | Release date: | 2006-06-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
1QDU
 
 | |
2CJY
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | Descriptor: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | Authors: | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | Deposit date: | 2006-04-09 | | Release date: | 2006-06-27 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
2CNO
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | Aza-peptide epoxide, Caspase-3 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Grutter, M.G. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
2CNL
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EPOXIDE, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Grutter, M.G, Powers, J.C. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
2CNK
 
 | | Crystal structures of caspase-3 in complex with aza-peptide epoxide inhibitors. | | Descriptor: | AZA-PEPTIDE EXPOXIDE, CASPASE-3 P12 SUBUNIT, CASPASE-3 P17 SUBUNIT | | Authors: | Ganesan, R, Jelakovic, S, Campbell, A.J, Li, Z.Z, Asgian, J.L, Powers, J.C, Grutter, M.G. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the S4 and S1 Prime Subsite Specificities in Caspase-3 with Aza-Peptide Epoxide Inhibitors
Biochemistry, 45, 2006
|
|
2C6Z
 
 | | crystal structure of dimethylarginine dimethylaminohydrolase I in complex with citrulline | | Descriptor: | CITRIC ACID, CITRULLINE, NG, ... | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2005-11-16 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
2CI6
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I bound with Zinc low pH | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ZINC ION | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inbitors
Structure, 14, 2006
|
|
2CI4
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I crystal form II | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CI7
 
 | | Crystal structure of Dimethylarginine Dimethylaminohydrolase I in complex with Zinc, high pH | | Descriptor: | GLYCINE, NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1, ... | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
