3EZB
 
 | | COMPLEX OF THE AMINO TERMINAL DOMAIN OF ENZYME I AND THE HISTIDINE-CONTAINING PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PHOSPHOCARRIER PROTEIN HPR), PROTEIN (PHOSPHOTRANSFER SYSTEM, ENZYME I) | | Authors: | Clore, G.M, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr.
Nat.Struct.Biol., 6, 1999
|
|
4Z8L
 
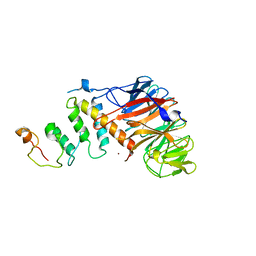 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
4HIR
 
 | |
1ATA
 
 | |
1ATB
 
 | |
1ATE
 
 | |
1ATD
 
 | |
5C8P
 
 | |
5C8O
 
 | |
5C8Q
 
 | | Crystal structure of MoCVNH3 variant (Mo0v) in complex with (N-GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MoCVNH3 variant | | Authors: | Koharudin, L.M.I, Gronenborn, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Fungal Cell Wall Recognition by a CVNH Protein with a Single LysM Domain.
Structure, 23, 2015
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
5GAT
 
 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
5HDW
 
 | |
2RMM
 
 | |
2RP3
 
 | |
2STW
 
 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
2STT
 
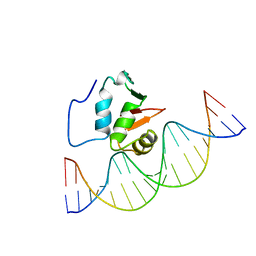 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, 25 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
1NEQ
 
 | |
1NER
 
 | |
1IL8
 
 | |
2HW0
 
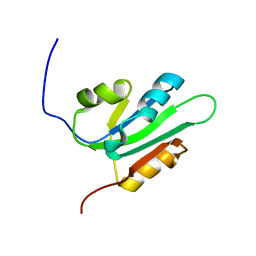 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
2HWT
 
 | |
3GAT
 
 | | SOLUTION NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, 34 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
1OLH
 
 | |
1OLG
 
 | |
