5LAL
 
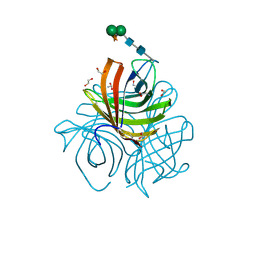 | | Structure of Arabidopsis dirigent protein AtDIR6 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dirigent protein 6, ... | | 著者 | Gasper, R, Kolesinski, P, Terlecka, B, Effenberger, I, Schaller, A, Hofmann, E. | | 登録日 | 2016-06-14 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6.
Plant Physiol., 172, 2016
|
|
5HI8
 
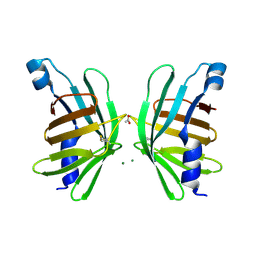 | | Structure of T-type Phycobiliprotein Lyase CpeT from Prochlorococcus phage P-HM1 | | 分子名称: | ACETATE ION, Antenna protein, MAGNESIUM ION | | 著者 | Gasper, R, Schwach, J, Frankenberg-Dinkel, N, Hofmann, E. | | 登録日 | 2016-01-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase Phi CpeT: THE ROLE OF AUXILIARY METABOLIC GENES.
J. Biol. Chem., 292, 2017
|
|
2HF9
 
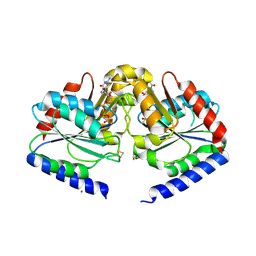 | | Crystal structure of HypB from Methanocaldococcus jannaschii in the triphosphate form | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Probable hydrogenase nickel incorporation protein hypB, ... | | 著者 | Gasper, R, Scrima, A, Wittinghofer, A. | | 登録日 | 2006-06-23 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into HypB, a GTP-binding protein that regulates metal binding.
J.Biol.Chem., 281, 2006
|
|
2HF8
 
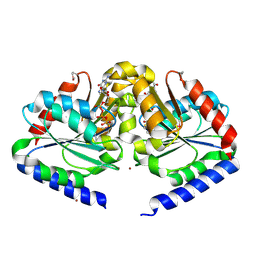 | | Crystal structure of HypB from Methanocaldococcus jannaschii in the triphosphate form, in complex with zinc | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Probable hydrogenase nickel incorporation protein hypB, ... | | 著者 | Gasper, R, Scrima, A, Wittinghofer, A. | | 登録日 | 2006-06-23 | | 公開日 | 2006-07-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into HypB, a GTP-binding protein that regulates metal binding.
J.Biol.Chem., 281, 2006
|
|
4TQ2
 
 | | Structure of S-type Phycobiliprotein Lyase CPES from Guillardia theta | | 分子名称: | HEXANE-1,6-DIOL, Putative phycoerythrin lyase | | 著者 | Gasper, R, Overkamp, K.E, Frankenberg-Dinkel, N, Hofmann, E. | | 登録日 | 2014-06-10 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Insights into the Biosynthesis and Assembly of Cryptophycean Phycobiliproteins.
J.Biol.Chem., 289, 2014
|
|
8ABX
 
 | | Crystal structure of IDO1 in complex with Apoxidole-1 | | 分子名称: | Indoleamine 2,3-dioxygenase 1, O1-tert-butyl O2-ethyl O5-methyl (E,5R)-5-(1-methylindol-2-yl)-5-[(4-methylphenyl)sulfonylamino]pent-2-ene-1,2,5-tricarboxylate, O2-tert-butyl O3-ethyl O6-methyl (2S,6R)-6-(1-methylindol-2-yl)-2,5-dihydro-1H-pyridine-2,3,6-tricarboxylate, ... | | 著者 | Dotsch, L, Ziegler, S, Waldmann, H, Gasper, R. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Identification of a Novel Pseudo-Natural Product Type IV IDO1 Inhibitor Chemotype.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8BWF
 
 | | PTBP1 RRM1 bound to an allosteric inhibitor | | 分子名称: | AMINO GROUP, GLYCEROL, Ligand, ... | | 著者 | Schmeing, S, Vetter, I, t Hart, P, Gasper, R. | | 登録日 | 2022-12-06 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rationally designed stapled peptides allosterically inhibit PTBP1-RNA-binding.
Chem Sci, 14, 2023
|
|
8C7I
 
 | |
5IXB
 
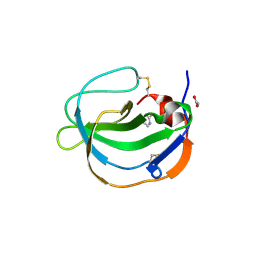 | | Structure of human Melanoma Inhibitory Activity (MIA) Protein in complex with Pyrimidin-2-amine | | 分子名称: | ACETATE ION, Melanoma-derived growth regulatory protein, PYRIMIDIN-2-AMINE | | 著者 | Yip, K.T, Gasper, R, Zhong, X.Y, Seibel, N, Puetz, S, Autzen, J, Scherkenbeck, J, Hofmann, E, Stoll, R. | | 登録日 | 2016-03-23 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Small Molecules Antagonise the MIA-Fibronectin Interaction in Malignant Melanoma.
Sci Rep, 6, 2016
|
|
7BOC
 
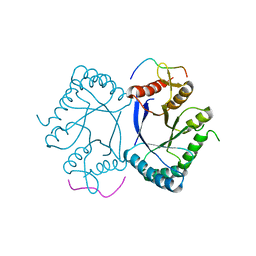 | | Crystal structure of the PRMT5 TIM barrel domain in complex with RioK1 peptide | | 分子名称: | Protein arginine N-methyltransferase 5, peptide | | 著者 | Krzyzanowski, A, t Hart, P, Waldmann, H, Gasper, R. | | 登録日 | 2021-01-25 | | 公開日 | 2021-09-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Biochemical Investigation of the Interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 Complex.
Chembiochem, 22, 2021
|
|
3BH7
 
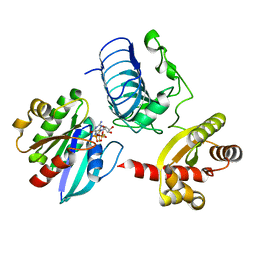 | | Crystal structure of the RP2-Arl3 complex bound to GDP-AlF4 | | 分子名称: | ADP-ribosylation factor-like protein 3, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Veltel, S, Gasper, R, Wittinghofer, A. | | 登録日 | 2007-11-28 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
3BH6
 
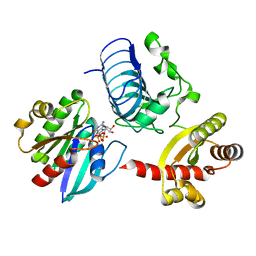 | | Crystal structure of the RP2-Arl3 complex bound to GppNHp | | 分子名称: | ADP-ribosylation factor-like protein 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Veltel, S, Gasper, R, Wittinghofer, A. | | 登録日 | 2007-11-28 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
5NS8
 
 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome in complex with inhibitor 1-Deoxynojirimycin | | 分子名称: | 1-DEOXYNOJIRIMYCIN, GLYCEROL, SULFATE ION, ... | | 著者 | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | 登録日 | 2017-04-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2019-01-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
5NS6
 
 | | Crystal structure of beta-glucosidase BglM-G1 from marine metagenome | | 分子名称: | Beta-glucosidase, GLYCEROL, SULFATE ION | | 著者 | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | 登録日 | 2017-04-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
5NS7
 
 | | Crystal structure of beta-glucosidase BglM-G1 mutant H75R from marine metagenome | | 分子名称: | GLYCEROL, SULFATE ION, beta-glucosidase M - G1 | | 著者 | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | 登録日 | 2017-04-25 | | 公開日 | 2018-08-08 | | 最終更新日 | 2019-01-30 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
6RMK
 
 | | Bacteriorhodopsin, dark state, cell 2, refined using the same protocol as sub-ps time delays | | 分子名称: | Bacteriorhodopsin, RETINAL | | 著者 | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | 登録日 | 2019-05-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6SBA
 
 | | Crystal Structure of mTEAD with a VGL4 Tertiary Structure Mimetic | | 分子名称: | ACETYL GROUP, GLYCEROL, Transcriptional enhancer factor TEF-3, ... | | 著者 | Adihou, H, Grossmann, T.N, Waldmann, H, Gasper, R. | | 登録日 | 2019-07-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A protein tertiary structure mimetic modulator of the Hippo signalling pathway.
Nat Commun, 11, 2020
|
|
6RXM
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16-Acetyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXQ
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after soaking | | 分子名称: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXK
 
 | | Crystal structure of CobB wt in complex with H4K16-Butyryl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | 分子名称: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXJ
 
 | | Crystal structure of CobB wt in complex with H4K16-Acetyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXP
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16-Crotonyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | 分子名称: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXR
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after co-crystallisation | | 分子名称: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Spinck, M, Gasper, R, Neumann, H. | | 登録日 | 2019-06-08 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
