7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 29_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWZ
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3K41
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M bound to Man-6-P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K43
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K42
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-dependent mannose-6-phosphate receptor, SN-GLYCEROL-1-PHOSPHATE, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3LHR
 
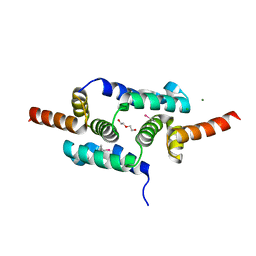 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
5UR5
 
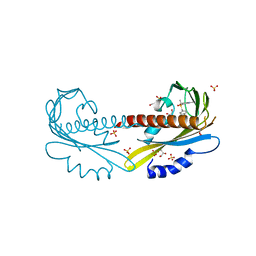 | | PYR1 bound to the rationally designed agonist 4m | | Descriptor: | Abscisic acid receptor PYR1, GLYCEROL, N-(4-cyano-3-ethyl-5-methylphenyl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5UR6
 
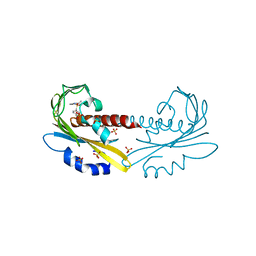 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5UR4
 
 | |
5UR7
 
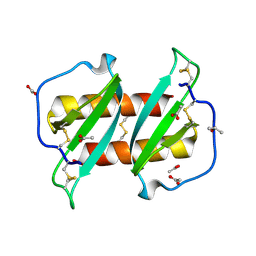 | |
5US5
 
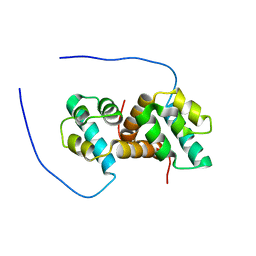 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
3NJ0
 
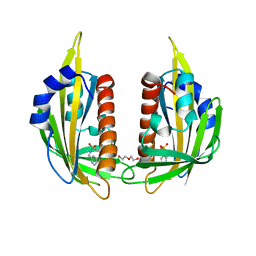 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJO
 
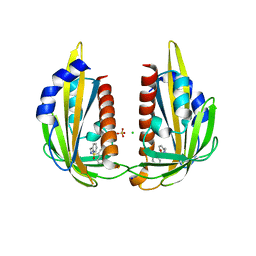 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5WKA
 
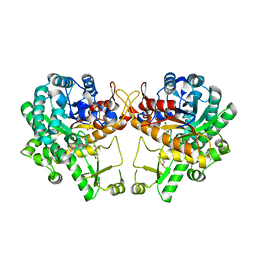 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
3NJ1
 
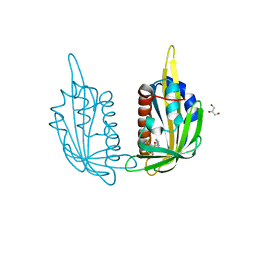 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6NWB
 
 | | PYL10 bound to the selective agonist hexabactin | | Descriptor: | Abscisic acid receptor PYL10, N-{[(4-cyanophenyl)methyl]sulfonyl}-1-(thiophen-3-yl)cyclohexane-1-carboxamide | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | PYL10 bound to the selective agonist hexabactin
To Be Published
|
|
6NWC
 
 | | PYL10 bound to the ABA pan-agonist 3CB | | Descriptor: | 1-{[(4-cyano-3-cyclopropylphenyl)acetyl]amino}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10 | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dynamic control of plant water use using designed ABA receptor agonists.
Science, 366, 2019
|
|
3DEU
 
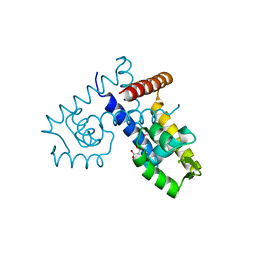 | | Crystal structure of transcription regulatory protein slyA from Salmonella typhimurium in complex with salicylate ligands | | Descriptor: | 2-HYDROXYBENZOIC ACID, Transcriptional regulator slyA | | Authors: | Le Trong, I, Brzovic, P.S, Fang, F.C, Libby, S.J, Stenkamp, R.E. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Evolution of SlyA/RovA Transcription Factors from Repressors to Countersilencers in Enterobacteriaceae .
Mbio, 10, 2019
|
|
2QED
 
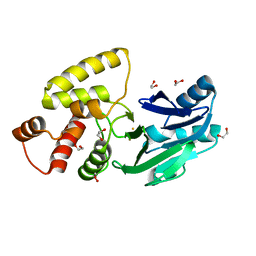 | | Crystal structure of Salmonella thyphimurium LT2 glyoxalase II | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Hydroxyacylglutathione hydrolase | | Authors: | Leite, N.R, Campos Bermudez, V.A, Krogh, R, Oliva, G, Soncini, F.C, Vila, A.J. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of Salmonella typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity
Biochemistry, 46, 2007
|
|
2ROC
 
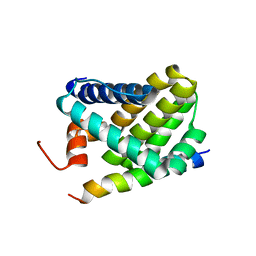 | | Solution structure of Mcl-1 Complexed with Puma | | Descriptor: | Bcl-2-binding component 3, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2REM
 
 | |
2ROD
 
 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
2RLJ
 
 | | NMR Structure of Ebola fusion peptide in SDS micelles at pH 7 | | Descriptor: | Envelope glycoprotein | | Authors: | Freitas, M.S, Gaspar, L.P, Lorenzoni, M, Almeida, F.C, Tinoco, L.W, Almeida, M.S, Maia, L.F, Degreve, L, Valente, A.P, Silva, J.L. | | Deposit date: | 2007-07-05 | | Release date: | 2007-08-07 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts.
J.Biol.Chem., 282, 2007
|
|
2V92
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
2V9J
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
