7LP5
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | Angiomotin,E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
7LP4
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
1XA6
 
 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1XAP
 
 | | Structure of the ligand binding domain of the Retinoic Acid Receptor beta | | Descriptor: | 4-[(1E)-2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PROP-1-ENYL]BENZOIC ACID, Retinoic acid receptor beta | | Authors: | Germain, P, Kammerer, S, Peluso-Iltis, C, Tortolani, D, Zusi, F.C, Starrett, J, Lapointe, P, Daris, J.P, Marinier, A, De Lera, A.R, Rochel, N, Gronemeyer, H. | | Deposit date: | 2004-08-26 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational design of RAR-selective ligands revealed by RARbeta crystal structure
Embo Rep., 5, 2004
|
|
1XFL
 
 | | Solution Structure of Thioredoxin h1 from Arabidopsis Thaliana | | Descriptor: | Thioredoxin h1 | | Authors: | Peterson, F.C, Lytle, B.L, Sampath, S, Vinarov, D, Tyler, E, Shahan, M, Markley, J.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-15 | | Release date: | 2004-09-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thioredoxin h1 from Arabidopsis thaliana.
Protein Sci., 14, 2005
|
|
7MLC
 
 | | PYL10 bound to the ABA pan-antagonist 4a | | Descriptor: | 1-{2-[3,5-dicyclopropyl-4-(4-{[(quinoxaline-2-carbonyl)amino]methyl}-1H-1,2,3-triazol-1-yl)phenyl]acetamido}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10, GLYCEROL | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MLD
 
 | | PYL10 bound to the ABA pan-antagonist antabactin | | Descriptor: | Abscisic acid receptor PYL10, CITRIC ACID, antabactin | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MWN
 
 | | An engineered PYL2-based WIN 55,212-2 synthetic cannabinoid sensor with a stabilized HAB1 variant | | Descriptor: | ACETATE ION, Abscisic acid receptor PYL2, CHLORIDE ION, ... | | Authors: | Peterson, F.C, Beltran, J, Bedewitz, M, Steiner, P.J, Cutler, S.R, Whitehead, T.A. | | Deposit date: | 2021-05-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Rapid biosensor development using plant hormone receptors as reprogrammable scaffolds.
Nat.Biotechnol., 40, 2022
|
|
1YEL
 
 | | Structure of the hypothetical Arabidopsis thaliana protein At1g16640.1 | | Descriptor: | At1g16640 | | Authors: | Peterson, F.C, Waltner, J.K, Lytle, B.L, Volkman, B.F, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the B3 domain from Arabidopsis thaliana protein At1g16640
Protein Sci., 14, 2005
|
|
1ZB8
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
1ZR9
 
 | |
2ADC
 
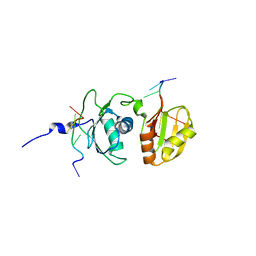 | | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
2AD9
 
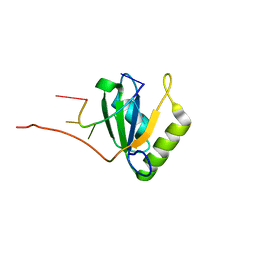 | | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
2A43
 
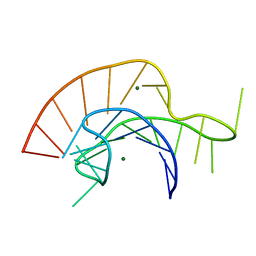 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
2ADB
 
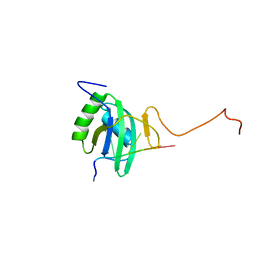 | | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
3QPZ
 
 | | Crystal structure of the N59A mutant of the 3-deoxy-d-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J, Cochrane, F.C. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TE5
 
 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TDH
 
 | | Structure of the regulatory fragment of sccharomyces cerevisiae AMPK in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
1PI1
 
 | | Crystal structure of a human Mob1 protein; toward understanding Mob-regulated cell cycle pathways. | | Descriptor: | Mob1A, ZINC ION | | Authors: | Stavridi, E.S, Harris, K.G, Huyen, Y, Bothos, J, Voewerd, P.M, Stayrook, S.E, Jeffrey, P.D, Pavletich, N.P, Luca, F.C. | | Deposit date: | 2003-05-29 | | Release date: | 2003-09-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human mob1 protein. Toward understanding mob-regulated cell cycle pathways.
Structure, 11, 2003
|
|
1Q53
 
 | |
1RJJ
 
 | | Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana | | Descriptor: | expressed protein | | Authors: | Cornilescu, G, Cornilescu, C.C, Zhao, Q, Frederick, R.O, Peterson, F.C, Thao, S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a homodimeric hypothetical protein, at5g22580, a structural genomics target from Arabidopsis thaliana.
J.Biomol.Nmr, 29, 2004
|
|
1RY4
 
 | |
1RZX
 
 | | Crystal Structure of a Par-6 PDZ-peptide Complex | | Descriptor: | Acetylated VKESLV Peptide, CG5884-PA | | Authors: | Peterson, F.C, Penkert, R.R, Volkman, F.B, Prehoda, K.E. | | Deposit date: | 2003-12-29 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition.
Mol.Cell, 13, 2004
|
|
