1TIH
 
 | |
2AJW
 
 | | Structure of the cyclic conotoxin MII-6 | | Descriptor: | Alpha-conotoxin MII | | Authors: | Clark, R.J, Fischer, H, Dempster, L, Daly, N.L, Rosengren, K.J, Nevin, S.T, Meunier, F.A, Adams, D.J, Craik, D.J. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the {alpha}-conotoxin MII.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2AK0
 
 | | Structure of cyclic conotoxin MII-7 | | Descriptor: | Alpha-conotoxin MII | | Authors: | Clark, R.J, Fischer, H, Dempster, L, Daly, N.L, Rosengren, K.J, Nevin, S.T, Meunier, F.A, Adams, D.J, Craik, D.J. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Engineering stable peptide toxins by means of backbone cyclization: Stabilization of the {alpha}-conotoxin MII.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2H8S
 
 | | Solution structure of alpha-conotoxin Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J, Fischer, H, Nevin, S.T, Adams, D.J, Craik, D.J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-06-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Synthesis, Structural Characterization, and Receptor Specificity of the {alpha}-Conotoxin Vc1.1.
J.Biol.Chem., 281, 2006
|
|
2H8B
 
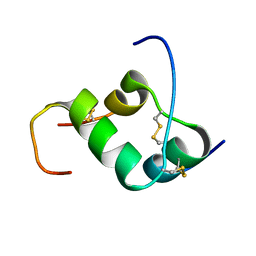 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
1IEN
 
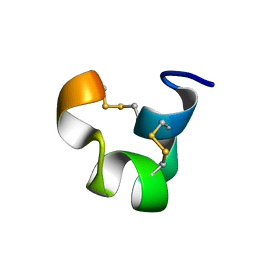 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1IEO
 
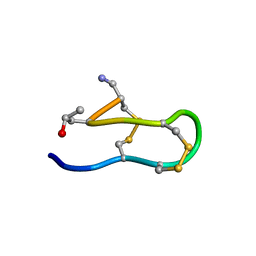 | | SOLUTION STRUCTURE OF MRIB-NH2 | | Descriptor: | PROTEIN MRIB-NH2 | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1R9I
 
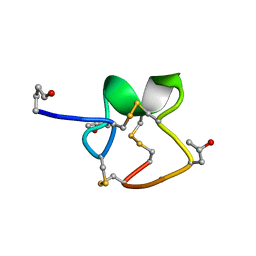 | | NMR Solution Structure of PIIIA toxin, NMR, 20 structures | | Descriptor: | Mu-conotoxin PIIIA | | Authors: | Nielsen, K.J, Watson, M, Adams, D.J, Hammarstrom, A.K, Gage, P.W, Hill, J.M, Craik, D.J, Thomas, L, Adams, D, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2003-10-30 | | Release date: | 2003-11-18 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mu-conotoxin PIIIA, a preferential inhibitor of persistent tetrodotoxin-sensitive sodium channels
J.Biol.Chem., 277, 2002
|
|
1RMK
 
 | | Solution structure of conotoxin MrVIB | | Descriptor: | Mu-O-conotoxin MrVIB | | Authors: | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
2PO8
 
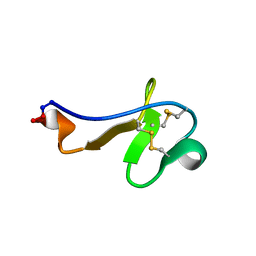 | |
1QH2
 
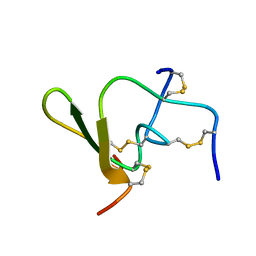 | |
6U7Q
 
 | | NMR solution structure of SFTI-R10 | | Descriptor: | GLY-ARG-CYS-THR-LYS-SER-ILE-PRO-PRO-ARG-CYS-PHE-PRO-ASP inhibitor | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U24
 
 | | NMR solution structure of triazole bridged SFTI-1 | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7U
 
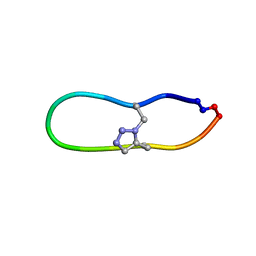 | | NMR solution structure of triazole bridged matriptase inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ARG-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7S
 
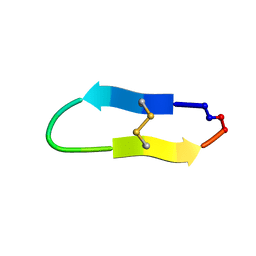 | |
6U7W
 
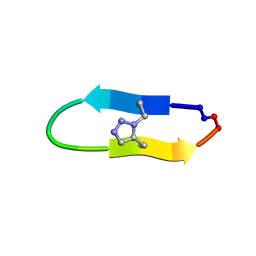 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U22
 
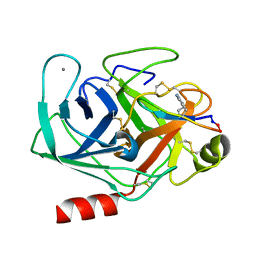 | | Crystal structure of SFTI-triazole inhibitor in complex with beta-trypsin | | Descriptor: | 1-methyl-1H-1,2,3-triazole, CALCIUM ION, Cationic trypsin, ... | | Authors: | White, A.M, King, G.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7X
 
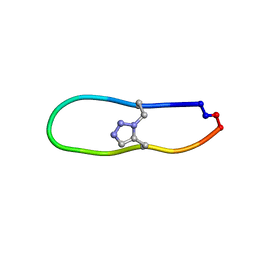 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7R
 
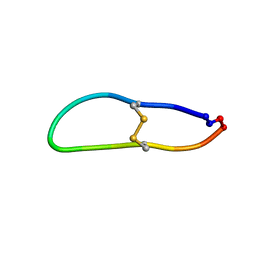 | |
6VH8
 
 | |
6VY8
 
 | |
6VXY
 
 | | Triazole bridged SFTI1 inhibitor in complex with beta-trypsin | | Descriptor: | 1-methyl-1H-1,2,3-triazole, CALCIUM ION, Cationic trypsin, ... | | Authors: | White, A.M, King, G.J, Durek, T, Craik, D.J. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 2020
|
|
5KWO
 
 | |
5KWX
 
 | |
5KWP
 
 | |
