4BA2
 
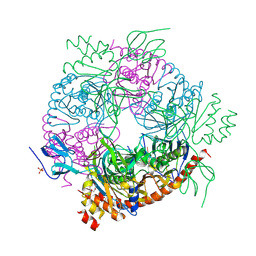 | |
4BA1
 
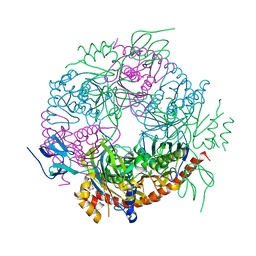 | |
1OL6
 
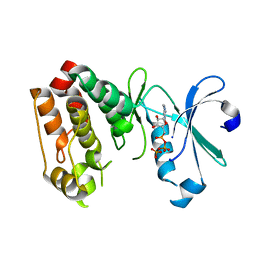 | |
1OL5
 
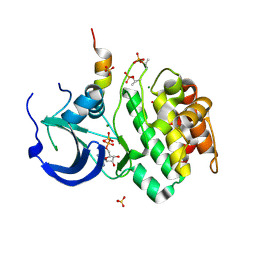 | | Structure of Aurora-A 122-403, phosphorylated on Thr287, Thr288 and bound to TPX2 1-43 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RESTRICTED EXPRESSION PROLIFERATION ASSOCIATED PROTEIN 100, ... | | Authors: | Bayliss, R, Conti, E. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Aurora-A Activation by Tpx2 at the Mitotic Spindle
Mol.Cell, 12, 2003
|
|
1OL7
 
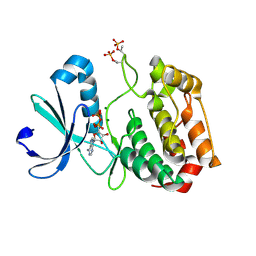 | |
1OF5
 
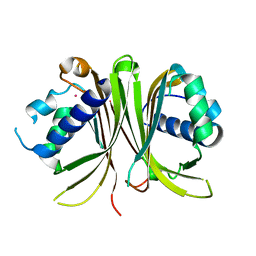 | | Crystal structure of Mex67-Mtr2 | | Descriptor: | MERCURY (II) ION, MRNA EXPORT FACTOR MEX67, MRNA TRANSPORT REGULATOR MTR2 | | Authors: | Fribourg, S, Conti, E. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15.
EMBO Rep., 4, 2003
|
|
5NKK
 
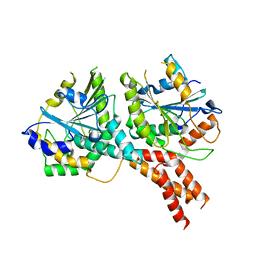 | | SMG8-SMG9 complex GDP bound | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
5NKM
 
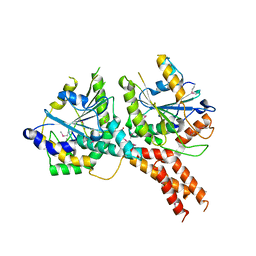 | | SMG8-SMG9 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
5OKZ
 
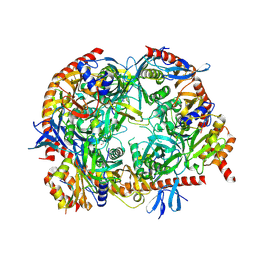 | | Crystal Strucrure of the Mpp6 Exosome complex | | Descriptor: | CHLORIDE ION, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Falk, S, Ebert, J, Conti, E. | | Deposit date: | 2017-07-26 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.200039 Å) | | Cite: | Mpp6 Incorporation in the Nuclear Exosome Contributes to RNA Channeling through the Mtr4 Helicase.
Cell Rep, 20, 2017
|
|
4AJ5
 
 | | Crystal structure of the Ska core complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 2, SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 3 | | Authors: | Jeyaprakash, A.A, Santamaria, A, Jayachandran, U, Chan, Y.W, Benda, C, Nigg, E.A, Conti, E. | | Deposit date: | 2012-02-15 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural and Functional Organization of the Ska Complex, a Key Component of the Kinetochore-Microtubule Interface.
Mol.Cell, 46, 2012
|
|
4A0J
 
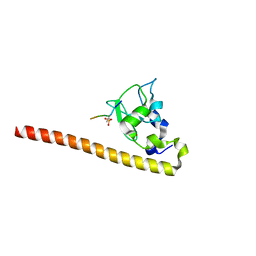 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A4K
 
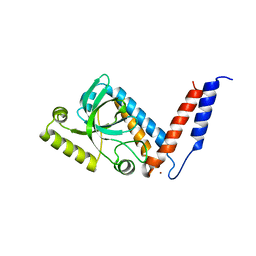 | |
4A6Q
 
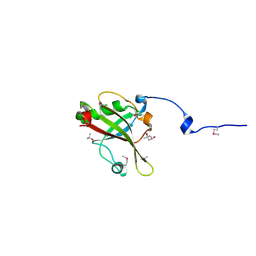 | | Crystal structure of mouse SAP18 residues 6-143 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, ISOPROPYL ALCOHOL | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-08 | | Release date: | 2012-03-07 | | Last modified: | 2015-03-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A4Z
 
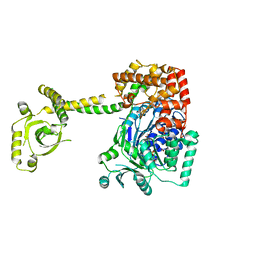 | | CRYSTAL STRUCTURE OF THE S. CEREVISIAE DEXH HELICASE SKI2 BOUND TO AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, ANTIVIRAL HELICASE SKI2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Halbach, F, Rode, M, Conti, E. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of S. Cerevisiae Ski2, a Dexh Helicase Associated with the Cytoplasmic Functions of the Exosome.
RNA, 18, 2012
|
|
4A0I
 
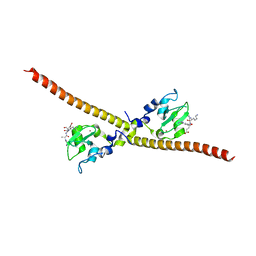 | | Crystal structure of Survivin bound to the N-terminal tail of hSgo1 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, SHUGOSHIN-LIKE 1, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0N
 
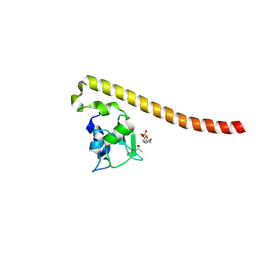 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A90
 
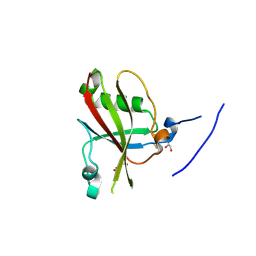 | | Crystal structure of mouse SAP18 residues 1-143 | | Descriptor: | GLYCEROL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18 | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8X
 
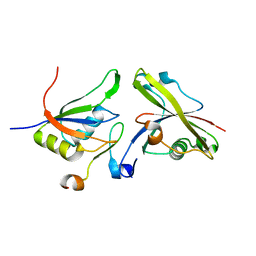 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4B8A
 
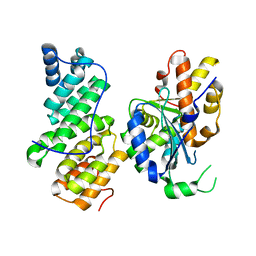 | | Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8B
 
 | | N-Terminal domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GOLD ION | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8C
 
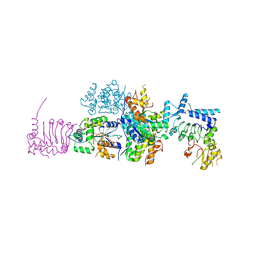 | | nuclease module of the yeast Ccr4-Not complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GLUCOSE-REPRESSIBLE ALCOHOL DEHYDROGENASE TRANSCRIPTIONAL EFFECTOR, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B89
 
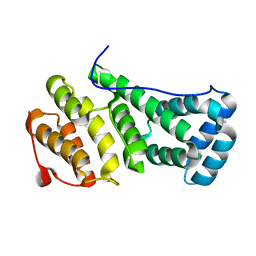 | | MIF4G domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4C9B
 
 | | Crystal structure of eIF4AIII-CWC22 complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Buchwald, G, Schuessler, S, Basquin, C, LeHir, H, Conti, E. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Eif4Aiii-Cwc22 Complex Shows How a Dead-Box Protein is Inhibited by a Mif4G Domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C92
 
 | | Crystal structure of the yeast Lsm1-7 complex | | Descriptor: | SM-LIKE PROTEIN LSM1, U6 SNRNA-ASSOCIATED SM-LIKE PROTEIN LSM2, U6 SNRNA-ASSOCIATED SM-LIKE PROTEIN LSM3, ... | | Authors: | Sharif, H, Conti, E. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic Mrna Turnover
Cell Rep., 5, 2013
|
|
4BRU
 
 | | Crystal structure of the yeast Dhh1-Edc3 complex | | Descriptor: | ATP-DEPENDENT RNA HELICASE DHH1, ENHANCER OF MRNA-DECAPPING PROTEIN 3 | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
