5AIE
 
 | | Not4 ring domain in complex with Ubc4 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 4, UBIQUITIN-CONJUGATING ENZYME E2 4, ZINC ION | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Last modified: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the Ubiquitylation Module of the Yeast Ccr4-not Complex.
Structure, 23, 2015
|
|
5AJD
 
 | |
5ANR
 
 | | Structure of a human 4E-T - DDX6 - CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TRANSPORTER, PROBABLE ATP-DEPENDENT RNA HELICASE DDX6 | | Authors: | Basquin, J, Oezguer, S, Conti, E. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of a Human 4E-T/Ddx6/Cnot1 Complex Reveals the Different Interplay of Ddx6-Binding Proteins with the Ccr4-not Complex.
Cell Rep., 13, 2015
|
|
5AOR
 
 | | Structure of MLE RNA ADP AlF4 complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP)-3', ADENOSINE-5'-DIPHOSPHATE, DOSAGE COMPENSATION REGULATOR, ... | | Authors: | Prabu, J.R, Conti, E. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the RNA Helicase Mle Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling.
Mol.Cell, 60, 2015
|
|
4BA2
 
 | |
4BA1
 
 | |
2J0B
 
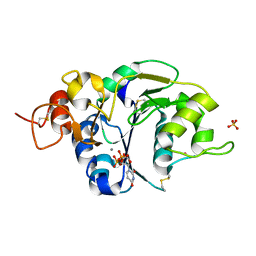 | | Structure of the catalytic domain of mouse Manic Fringe in complex with UDP and manganese | | Descriptor: | BETA-1,3-N-ACETYLGLUCOSAMINYLTRANSFERASE MANIC FRINGE, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jinek, M, Chen, Y.-W, Clausen, H, Cohen, S.M, Conti, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-04 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Notch-Modifying Glycosyltransferase Fringe
Nat.Struct.Mol.Biol., 13, 2006
|
|
2J0A
 
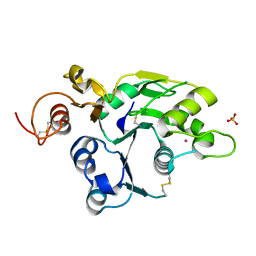 | | Structure of the catalytic domain of mouse Manic Fringe | | Descriptor: | BETA-1,3-N-ACETYLGLUCOSAMINYLTRANSFERASE MANIC FRINGE, POTASSIUM ION, SULFATE ION | | Authors: | Jinek, M, Chen, Y.-W, Clausen, H, Cohen, S.M, Conti, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Notch-Modifying Glycosyltransferase Fringe
Nat.Struct.Mol.Biol., 13, 2006
|
|
4A90
 
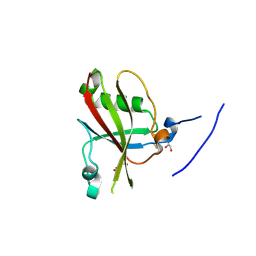 | | Crystal structure of mouse SAP18 residues 1-143 | | Descriptor: | GLYCEROL, HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18 | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8X
 
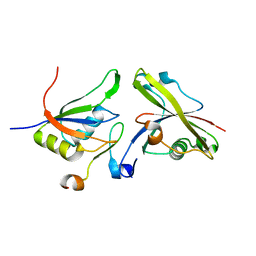 | | Structure of the core ASAP complex | | Descriptor: | HISTONE DEACETYLASE COMPLEX SUBUNIT SAP18, HOOK-LIKE, ISOFORM A, ... | | Authors: | Murachelli, A.G, Ebert, J, Basquin, C, Le Hir, H, Conti, E. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of the Asap Core Complex Reveals the Existence of a Pinin-Containing Psap Complex
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A0I
 
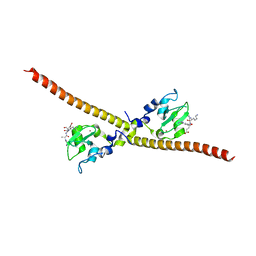 | | Crystal structure of Survivin bound to the N-terminal tail of hSgo1 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, SHUGOSHIN-LIKE 1, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0J
 
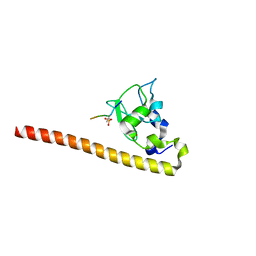 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4A0N
 
 | | Crystal structure of Survivin bound to the phosphorylated N-terminal tail of histone H3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, HISTONE H3 PEPTIDE, ZINC ION | | Authors: | Jeyaprakash, A.A, Basquin, C, Jayachandran, U, Conti, E. | | Deposit date: | 2011-09-09 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structural Basis for the Recognition of Phosphorylated Histone H3 by the Survivin Subunit of the Chromosomal Passenger Complex.
Structure, 19, 2011
|
|
4B8B
 
 | | N-Terminal domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GOLD ION | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B89
 
 | | MIF4G domain of the yeast Not1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8C
 
 | | nuclease module of the yeast Ccr4-Not complex | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, GLUCOSE-REPRESSIBLE ALCOHOL DEHYDROGENASE TRANSCRIPTIONAL EFFECTOR, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4B8A
 
 | | Structure of yeast NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 1, POLY(A) RIBONUCLEASE POP2 | | Authors: | Basquin, J, Conti, E. | | Deposit date: | 2012-08-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the Nuclease Module of the Yeast Ccr4-not Complex: The not1-Caf1-Ccr4 Interaction.
Mol.Cell, 48, 2012
|
|
4C9B
 
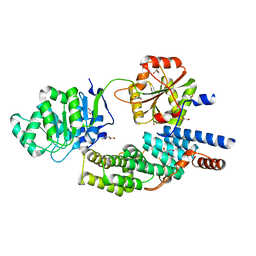 | | Crystal structure of eIF4AIII-CWC22 complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Buchwald, G, Schuessler, S, Basquin, C, LeHir, H, Conti, E. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Eif4Aiii-Cwc22 Complex Shows How a Dead-Box Protein is Inhibited by a Mif4G Domain
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BY6
 
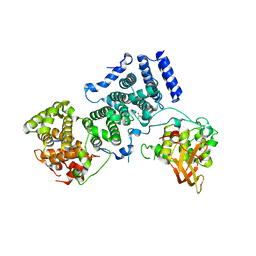 | | Yeast Not1-Not2-Not5 complex | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-16 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structure and RNA-Binding Properties of the not1-not2-not5 Module of the Yeast Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BUJ
 
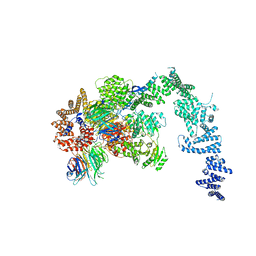 | | Crystal structure of the S. cerevisiae Ski2-3-8 complex | | Descriptor: | ANTIVIRAL HELICASE SKI2, ANTIVIRAL PROTEIN SKI8, SULFATE ION, ... | | Authors: | Halbach, F, Reichelt, P, Rode, M, Conti, E. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The Yeast Ski Complex: Crystal Structure and RNA Channeling to the Exosome Complex.
Cell(Cambridge,Mass.), 154, 2013
|
|
4C92
 
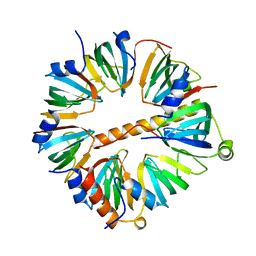 | | Crystal structure of the yeast Lsm1-7 complex | | Descriptor: | SM-LIKE PROTEIN LSM1, U6 SNRNA-ASSOCIATED SM-LIKE PROTEIN LSM2, U6 SNRNA-ASSOCIATED SM-LIKE PROTEIN LSM3, ... | | Authors: | Sharif, H, Conti, E. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic Mrna Turnover
Cell Rep., 5, 2013
|
|
4BRW
 
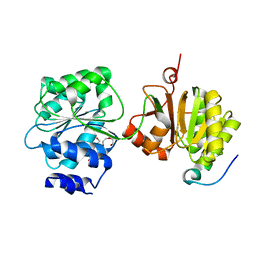 | | Crystal structure of the yeast Dhh1-Pat1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-DEPENDENT RNA HELICASE DHH1, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, ... | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
4BRU
 
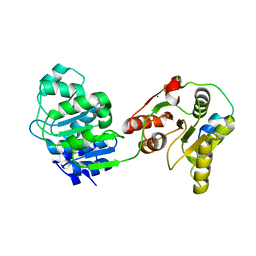 | | Crystal structure of the yeast Dhh1-Edc3 complex | | Descriptor: | ATP-DEPENDENT RNA HELICASE DHH1, ENHANCER OF MRNA-DECAPPING PROTEIN 3 | | Authors: | Sharif, H, Ozgur, S, Sharma, K, Basquin, C, Urlaub, H, Conti, E. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Structural Analysis of the Yeast Dhh1-Pat1 Complex Reveals How Dhh1 Engages Pat1, Edc3 and RNA in Mutually Exclusive Interactions
Nucleic Acids Res., 41, 2013
|
|
4C8Q
 
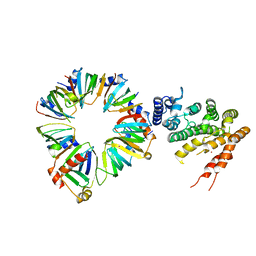 | | Crystal structure of the yeast Lsm1-7-Pat1 complex | | Descriptor: | COBALT (II) ION, DNA TOPOISOMERASE 2-ASSOCIATED PROTEIN PAT1, SM-LIKE PROTEIN LSM1, ... | | Authors: | Sharif, H, Conti, E. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Architecture of the Lsm1-7-Pat1 Complex: A Conserved Assembly in Eukaryotic Mrna Turnover
Cell Rep., 5, 2013
|
|
4CT6
 
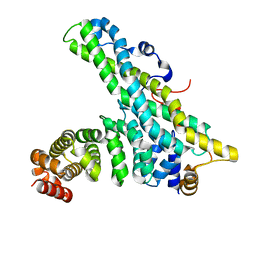 | | CNOT9-CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
