2L1F
 
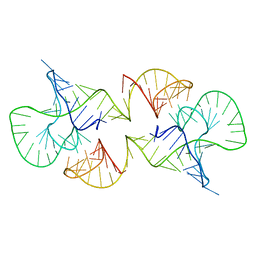 | | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography | | Descriptor: | RNA (65-MER), RNA (66-MER) | | Authors: | Summers, M.F, Irobalieva, R.N, Tolbert, B, Smalls-Manty, A, Iyalla, K, Loeliger, K, D'Souza, V, Khant, H, Schmid, M, Garcia, E, Telesnitsky, A, Chiu, W, Miyazaki, Y. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-27 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.
J.Mol.Biol., 404, 2010
|
|
1TT9
 
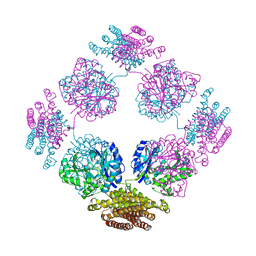 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
3J03
 
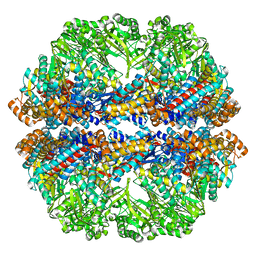 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
3KTT
 
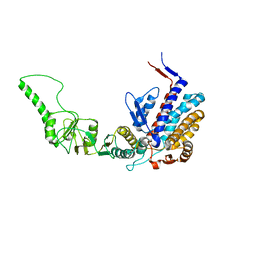 | | Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit beta | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-25 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7JM3
 
 | | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer | | Descriptor: | Matrix protein 1 | | Authors: | Su, Z, Pintilie, G, Selzer, L, Chiu, W, Kirkegaard, K. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Full-length three-dimensional structure of the influenza A virus M1 protein and its organization into a matrix layer.
Plos Biol., 18, 2020
|
|
6UKT
 
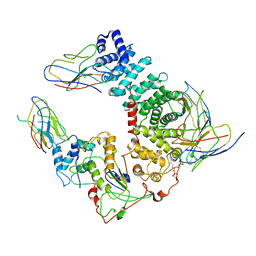 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | Authors: | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
6UES
 
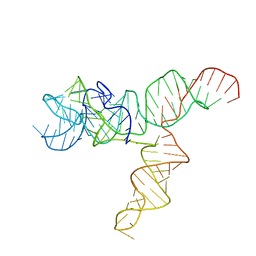 | | Apo SAM-IV Riboswitch | | Descriptor: | RNA (119-MER) | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UET
 
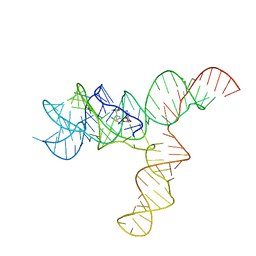 | | SAM-bound SAM-IV riboswitch | | Descriptor: | RNA (119-MER), S-ADENOSYLMETHIONINE | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6V5C
 
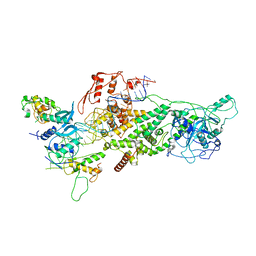 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6V5B
 
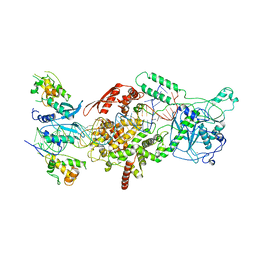 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6VQV
 
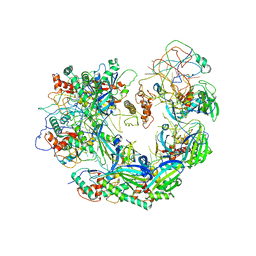 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
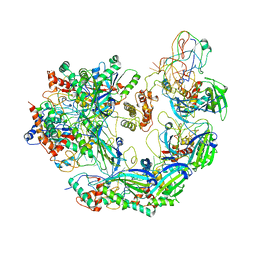 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
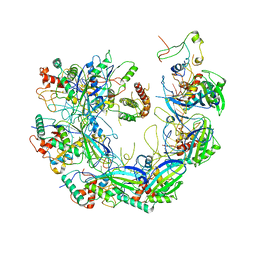 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
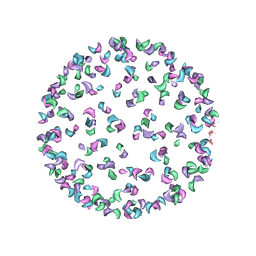
3J0G
 
 | |
3J0C
 
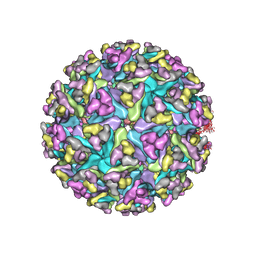 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
7EZ0
 
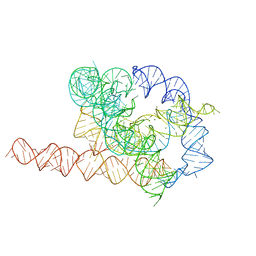 | | Apo L-21 ScaI Tetrahymena ribozyme | | Descriptor: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
7EZ2
 
 | | Holo L-16 ScaI Tetrahymena ribozyme | | Descriptor: | Holo L-16 ScaI Tetrahymena ribozyme, Holo L-16 ScaI Tetrahymena ribozyme S1, Holo L-16 ScaI Tetrahymena ribozyme S2, ... | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
3IZN
 
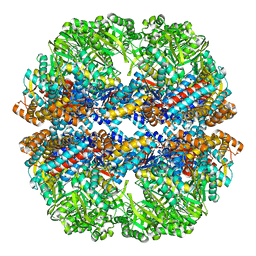 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
7K3V
 
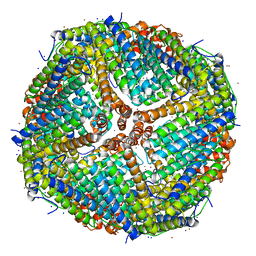 | | Apoferritin structure at 1.34 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with K3 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.34 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7K3W
 
 | | Apoferritin structure at 1.36 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with Falcon4 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.36 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
7KIP
 
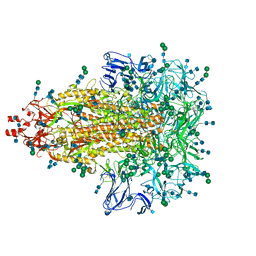 | | A 3.4 Angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Chmielewski, D, Schmid, M, Simmons, G, Jin, J, Chiu, W. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A 3.4- angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles.
Biorxiv, 2020
|
|
3IXV
 
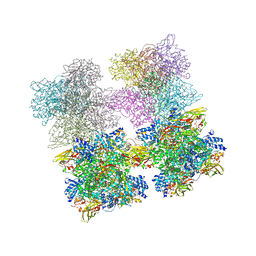 | | Scorpion Hemocyanin resting state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3IZI
 
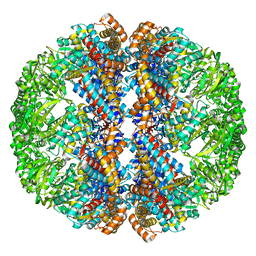 | | Mm-cpn rls with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IXW
 
 | | Scorpion Hemocyanin activated state pseudo atomic model built based on cryo-EM density map | | Descriptor: | Hemocyanin AA6 chain | | Authors: | Cong, Y, Zhang, Q, Woolford, D, Schweikardt, T, Khant, H, Ludtke, S, Chiu, W, Decker, H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural Mechanism of SDS-Induced Enzyme Activity of Scorpion Hemocyanin Revealed by Electron Cryomicroscopy.
Structure, 17, 2009
|
|
3IZJ
 
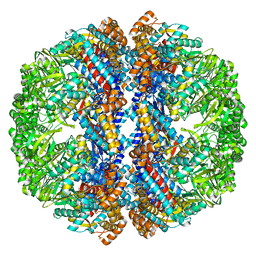 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
