5IG7
 
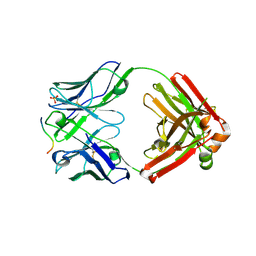 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment in complex of peptide PLQPQQPFP | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-02-27 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
5IK3
 
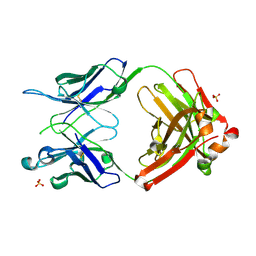 | | Crystal structure of anti-gliadin 1002-1E03 Fab fragment | | Descriptor: | 1E03 Fab fragment heavy chain, 1E03 Fab fragment light chain, SULFATE ION | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
5IJK
 
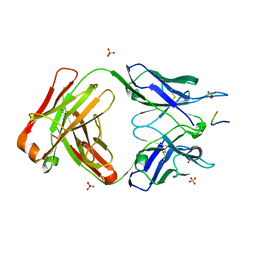 | | Crystal structure of anti-gliadin 1002-1E03 Fab fragment in complex of peptide PLQPEQPFP | | Descriptor: | 1E03 Fab fragment heavy chain, 1E03 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
5IFJ
 
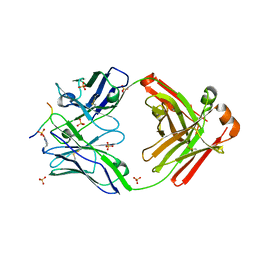 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment in complex of peptide PLQPEQPFP | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
4IAN
 
 | | Crystal Structure of apo Human PRPF4B kinase domain | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-06 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IFC
 
 | | Crystal Structure of ADP-bound Human PRPF4B kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-14 | | Release date: | 2013-08-28 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IJP
 
 | | Crystal Structure of Human PRPF4B kinase domain in complex with 4-{5-[(2-Chloro-pyridin-4-ylmethyl)-carbamoyl]-thiophen-2-yl}-benzo[b]thiophene-2-carboxylic acid amine | | Descriptor: | 4-(5-{[(2-chloropyridin-4-yl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4ICK
 
 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4IJX
 
 | |
4IIR
 
 | | Crystal Structure of AMPPNP-bound Human PRPF4B kinase domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
7ET0
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB from wPip | | Descriptor: | Bacteria factor A, Bacteria factor B | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESX
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wPip | | Descriptor: | Bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESZ
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB with Mn2+ from wPip | | Descriptor: | BACTERIA FACTOR A, BACTERIA FACTOR B, MANGANESE (II) ION | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESY
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip | | Descriptor: | Bacteria factor 1, CALCIUM ION, ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6L03
 
 | | structure of PTP-MEG2 and MUNC18-1-pY145 peptide complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 9, stxbp1-pY145 peptide | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
6KZQ
 
 | | structure of PTP-MEG2 and NSF-pY83 peptide complex | | Descriptor: | NSF-pY83 peptide, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Chen, X, Yu, X, Sun, J.P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PTP-MEG2 regulates quantal size and fusion pore opening through two distinct structural bases and substrates.
Embo Rep., 22, 2021
|
|
4NKJ
 
 | |
5Z9X
 
 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5Z9Z
 
 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R83
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R9V
 
 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R36
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Ngo, A, Fong, K, Cox, D, Fisher, A, Chen, X. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R84
 
 | | Crystal structure of Sialyltransferase from Photobacterium damsela with CMP-3F(a)Neu5Ac bound | | Descriptor: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
