4WIS
 
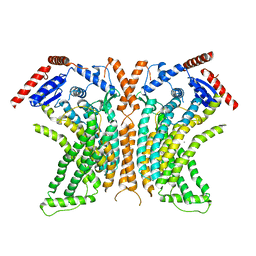 | | Crystal structure of the lipid scramblase nhTMEM16 in crystal form 1 | | Descriptor: | CALCIUM ION, lipid scramblase | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
1IG8
 
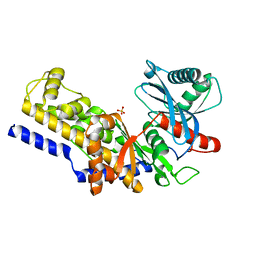 | | Crystal Structure of Yeast Hexokinase PII with the correct amino acid sequence | | Descriptor: | SULFATE ION, hexokinase PII | | Authors: | Kuser, P.R, Krauchenco, S, Antunes, O.A, Polikarpov, I. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action.
J.Biol.Chem., 275, 2000
|
|
7TOC
 
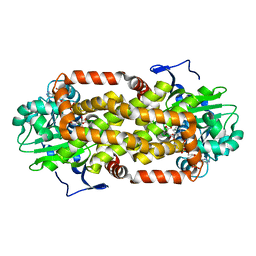 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
6YVG
 
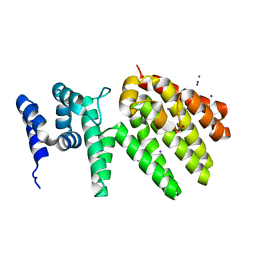 | | Crystal structure of MesI (Lpg2505) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MesI (Lpg2505) | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the metaeffector MesI (Lpg2505) from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
4IM6
 
 | | LRR domain from human NLRP1 | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hahne, G, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2013-01-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the leucine-rich repeat domain of the NOD-like receptor NLRP1: implications for binding of muramyl dipeptide.
Febs Lett., 588, 2014
|
|
8BVP
 
 | | Crystal structure of an N-terminal fragment of the effector protein Lpg2504 (SidI) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Restriction endonuclease | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal domain of the effector protein SidI of Legionella pneumophila reveals a glucosyl transferase domain.
Biochem.Biophys.Res.Commun., 661, 2023
|
|
5M2D
 
 | |
5MRJ
 
 | | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum | | Descriptor: | Beta-xylanase, SULFATE ION | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum
To Be Published
|
|
5MRS
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF
To Be Published
|
|
5MRR
 
 | | Crystal structure of L1 protease of Lysobacter sp. XL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of L1 protease of Lysobacter sp. XL1
To Be Published
|
|
8CRV
 
 | | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Carbamate kinase, FORMIC ACID, ... | | Authors: | Kim, Y, Skarina, T, Mesa, N, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa
To Be Published
|
|
5MRT
 
 | | Crystal structure of L5 protease Lysobacter sp. XL1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of L5 protease Lysobacter sp. XL1
To Be Published
|
|
5M6G
 
 | | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea | | Descriptor: | Beta-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Lisov, A, Leontievsky, A. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Crystal structure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea
To Be Published
|
|
5M0K
 
 | |
2AKA
 
 | | Structure of the nucleotide-free myosin II motor domain from Dictyostelium discoideum fused to the GTPase domain of dynamin 1 from Rattus norvegicus | | Descriptor: | Dynamin-1, LINKER, myosin II heavy chain | | Authors: | Reubold, T.F, Eschenburg, S, Becker, A, Leonard, M, Schmid, S.L, Vallee, R.B, Kull, F.J, Manstein, D.J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GTPase domain of rat dynamin 1.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1G6T
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G6S
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE AND GLYPHOSATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, GLYPHOSATE, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2MLL
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
1H0P
 
 | | Cyclophilin_5 from C. elegans | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 5 | | Authors: | Picken, N.C, Eschenlauer, S, Taylor, P, Page, A.P, Walkinshaw, M.D. | | Deposit date: | 2002-06-26 | | Release date: | 2002-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biological Characterisation of the Gut-Associated Cyclophilin B Isoforms from Caenorhabditis Elegans
J.Mol.Biol., 322, 2002
|
|
8Q7Y
 
 | |
8QDO
 
 | | Crystal structure of the tegument protein UL82 (pp71) from Human Cytomegalovirus | | Descriptor: | Protein pp71, TETRAETHYLENE GLYCOL | | Authors: | Bresch, I.P, Eberhage, J, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tegument protein UL82 (pp71) from human cytomegalovirus.
Protein Sci., 33, 2024
|
|
2WVW
 
 | | Cryo-EM structure of the RbcL-RbcX complex | | Descriptor: | RBCX PROTEIN, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN | | Authors: | Liu, C, Young, A.L, Starling-Windhof, A, Bracher, A, Saschenbrecker, S, Rao, B.V, Rao, K.V, Berninghausen, O, Mielke, T, Hartl, F.U, Beckmann, R, Hayer-Hartl, M. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Coupled Chaperone Action in Folding and Assembly of Hexadecameric Rubisco
Nature, 463, 2010
|
|
1L3P
 
 | | CRYSTAL STRUCTURE OF THE FUNCTIONAL DOMAIN OF THE MAJOR GRASS POLLEN ALLERGEN Phl p 5b | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, POLLEN ALLERGEN Phl p 5b | | Authors: | Rajashankar, K.R, Bufe, A, Weber, W, Eschenburg, S, Lindner, B, Betzel, C. | | Deposit date: | 2002-02-28 | | Release date: | 2003-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the functional domain of the major grass-pollen allergen Phlp 5b.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6IEW
 
 | |
1W70
 
 | | SH3 domain of p40phox complexed with C-terminal polyProline region of p47phox | | Descriptor: | NEUTROPHIL CYTOSOL FACTOR 1, NEUTROPHIL CYTOSOL FACTOR 4, SULFATE ION, ... | | Authors: | Massenet, C, Chenavas, S, Cohen-Addad, C, Dagher, M.-C, Brandolin, G, Pebay-Peyroula, E, Fieschi, F. | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Effects of P47Phox C-Terminus Phosphorylation on Binding Interactions with P40Phox and P67Phox: Structural and Functional Comparison of P40Phox P67Phox SH3 Domains
J.Biol.Chem., 280, 2005
|
|
