1OV4
 
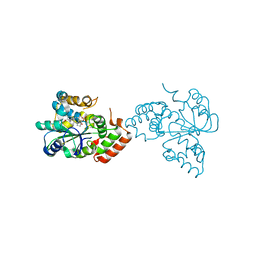 | | Crystal structure of human DHEA-ST complexed with androsterone | | 分子名称: | (3Beta,5alpha)-3-Hydroxyandrostan-17-one, Alcohol sulfotransferase, SULFATE ION | | 著者 | Chang, H.J, Shi, R, Rhese, P, Lin, S.X. | | 登録日 | 2003-03-25 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex.
J.Biol.Chem., 279, 2004
|
|
3AUV
 
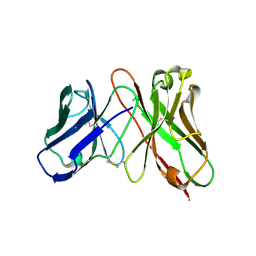 | | Predicting Amino Acid Preferences in the Complementarity Determining Regions of an Antibody-Antigen Recognition Interface | | 分子名称: | sc-dsFv derived from the G6-Fab | | 著者 | Yu, C.M, Peng, H.P, Chen, I.C, Lee, Y.C, Chen, J.B, Tsai, K.C, Chen, C.T, Chang, J.Y, Yang, E.W, Hsu, P.C, Jian, J.W, Hsu, H.J, Chang, H.J, Hsu, W.L, Huang, K.F, Ma, A.C, Yang, A.S. | | 登録日 | 2011-02-16 | | 公開日 | 2012-02-22 | | 最終更新日 | 2012-04-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rationalization and design of the complementarity determining region sequences in an antibody-antigen recognition interface
Plos One, 7, 2012
|
|
3PJF
 
 | |
3PJE
 
 | |
3PJD
 
 | |
3PG4
 
 | |
4DIT
 
 | |
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | 分子名称: | MONELLIN | | 著者 | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | 登録日 | 1998-08-06 | | 公開日 | 1999-06-08 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | 分子名称: | MONELLIN | | 著者 | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | 登録日 | 2000-09-18 | | 公開日 | 2001-06-06 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
3V9B
 
 | |
4KBZ
 
 | |
