5LV2
 
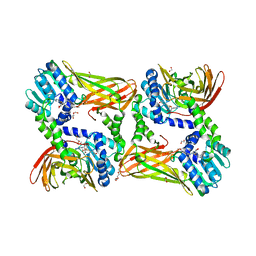 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1246 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
5LV5
 
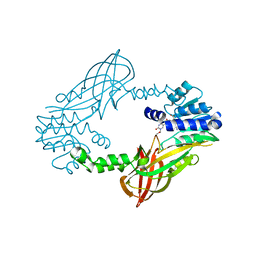 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
5MFT
 
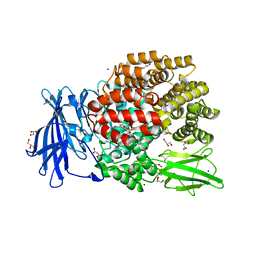 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-1-bromo-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFR
 
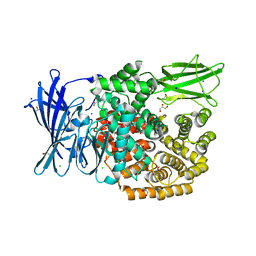 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFS
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
4C4A
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SAH | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, PROTEIN ARGININE N-METHYLTRANSFERASE 7, ... | | Authors: | Cura, V, Troffer-Charlier, N, Bonnefond, L, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-09-02 | | Release date: | 2014-07-02 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight Into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State Into a Single Active Site.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1OHF
 
 | | The refined structure of Nudaurelia capensis omega virus | | Descriptor: | MAGNESIUM ION, NUDAURELIA CAPENSIS OMEGA VIRUS CAPSID PROTEIN | | Authors: | Helgstrand, C, Munshi, S, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2004-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Refined Structure of Nudaurelia Capensis Omega Virus Reveals Control Elements for a T = 4 Capsid Maturation
Virology, 318, 2004
|
|
4BZ8
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1038 | | Descriptor: | (2R)-2-methyl-3-oxo-4H-1,4-benzothiazine-6-carbohydroxamic acid, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ6
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with SAHA | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ9
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1075 | | Descriptor: | 3-chlorobenzothiophene-2-carbohydroxamic acid, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ5
 
 | | Crystal structure of Schistosoma mansoni HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ7
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4CQF
 
 | |
