5IM4
 
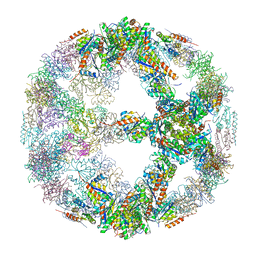 | | Crystal structure of designed two-component self-assembling icosahedral cage I52-32 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, Phosphotransferase system, mannose/fructose-specific component IIA | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
1K26
 
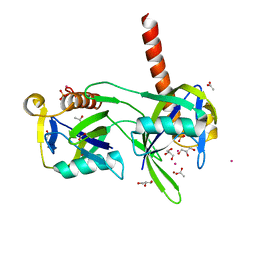 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6P0U
 
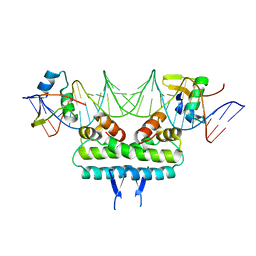 | | Crystal structure of ternary DNA complex " FX(1-2)-2Xis" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
1K2E
 
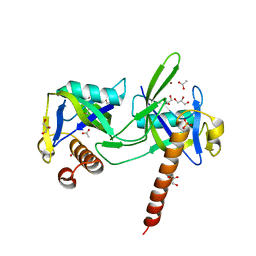 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5K7P
 
 | | MicroED structure of xylanase at 2.3 A resolution | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
6PIL
 
 | | Antibody scFv-M204 monomeric state | | Descriptor: | CHLORIDE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
6P0S
 
 | | Crystal structure of ternary DNA complex "FX2" containing E. coli Fis and phage lambda Xis | | Descriptor: | DNA (27-MER), FX1-2, DNA-binding protein Fis, ... | | Authors: | Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2019-05-17 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative DNA binding by proteins through DNA shape complementarity.
Nucleic Acids Res., 47, 2019
|
|
5K7R
 
 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
1JRK
 
 | | Crystal Structure of a Nudix Protein from Pyrobaculum aerophilum Reveals a Dimer with Intertwined Beta Sheets | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nudix homolog | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6PK8
 
 | | Antibody scFv-M204 dimeric state | | Descriptor: | SULFATE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
5K7Q
 
 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7O
 
 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7S
 
 | | MicroED structure of proteinase K at 1.6 A resolution | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
6PSC
 
 | | Antibody scFv-M204 trimeric state | | Descriptor: | scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-07-12 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
6M7M
 
 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
1L9L
 
 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
1LMI
 
 | | 1.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED PROTEIN FROM MYCOBACTERIUM TUBERCULOSIS-MPT63 | | Descriptor: | Immunogenic protein MPT63/MPB63 | | Authors: | Goulding, C.W, Parseghian, A, Sawaya, M.R, Cascio, D, Apostol, M, Gennaro, M.L, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-01 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a major secreted protein of Mycobacterium tuberculosis-MPT63 at
1.5-A resolution
Protein Sci., 11, 2002
|
|
7LUZ
 
 | |
7LV2
 
 | |
7MPK
 
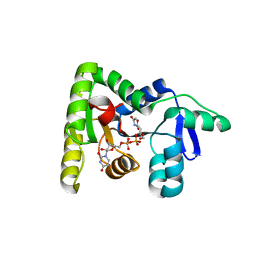 | | Crystal structure of TagA with UDP-GlcNAc | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
7N41
 
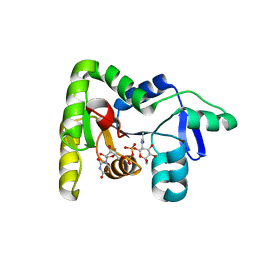 | | Crystal structure of TagA with UDP-ManNAc | | Descriptor: | (2R,3S,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-06-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
307D
 
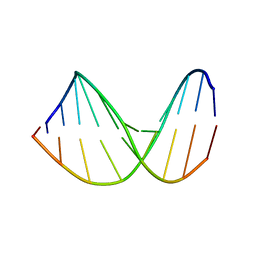 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
1YAC
 
 | |
