7TFA
 
 | | P. polymyxa GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TF7
 
 | | S. aureus GS(12) - apo | | Descriptor: | Glutamine synthetase | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TEN
 
 | |
7TF6
 
 | | S. aureus GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFB
 
 | | P. polymyxa GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
3BR3
 
 | |
3BR1
 
 | |
3BR6
 
 | |
3BR5
 
 | |
3BR2
 
 | |
3BR0
 
 | |
3BQZ
 
 | |
3DNU
 
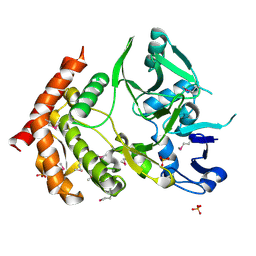 | | structure of MDT protein | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein hipA | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DNT
 
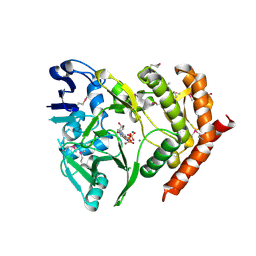 | | structures of MDT proteins | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein hipA, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3FBR
 
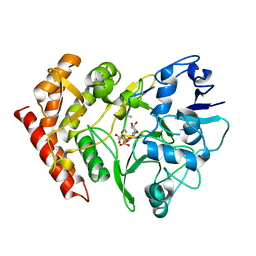 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3GCT
 
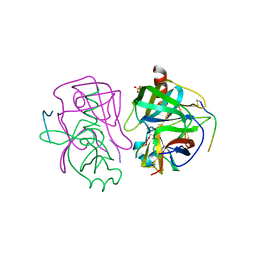 | |
5CRO
 
 | |
1QB8
 
 | |
1HK9
 
 | |
1KNX
 
 | | HPr kinase/phosphatase from Mycoplasma pneumoniae | | Descriptor: | Probable HPr(Ser) kinase/phosphatase | | Authors: | Allen, G.S. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HPr Kinase/Phosphatase from Mycoplasma pneumoniae
J.Mol.Biol., 326, 2003
|
|
1LBG
 
 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBH
 
 | | INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, INTACT LACTOSE OPERON REPRESSOR WITH GRATUITOUS INDUCER IPTG | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1LBI
 
 | | LAC REPRESSOR | | Descriptor: | LAC REPRESSOR | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-02-17 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
1GCT
 
 | |
