1HL9
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH A MECHANISM BASED INHIBITOR | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, PUTATIVE ALPHA-L-FUCOSIDASE | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
1HM9
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A AND UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, ... | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
1I12
 
 | |
1I1D
 
 | | CRYSTAL STRUCTURE OF YEAST GNA1 BOUND TO COA AND GLNAC-6P | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, GLUCOSAMINE-PHOSPHATE N-ACETYLTRANSFERASE, ... | | Authors: | Peneff, C, Mengin-Lecreulx, D, Bourne, Y. | | Deposit date: | 2001-02-01 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase.
J.Biol.Chem., 276, 2001
|
|
1I21
 
 | | CRYSTAL STRUCTURE OF YEAST GNA1 | | Descriptor: | GLUCOSAMINE-PHOSPHATE N-ACETYLTRANSFERASE | | Authors: | Peneff, C, Mengin-Lecreulx, D, Bourne, Y. | | Deposit date: | 2001-02-05 | | Release date: | 2001-05-16 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase.
J.Biol.Chem., 276, 2001
|
|
2WQZ
 
 | | Crystal structure of synaptic protein neuroligin-4 in complex with neurexin-beta 1: alternative refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEUREXIN-1-BETA, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and its Beta-Neurexin Complex: Determinants for Folding and Cell Adhesion.
Neuron, 56, 2007
|
|
2WNJ
 
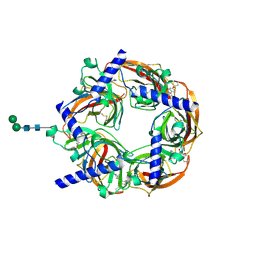 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH DMXBA | | Descriptor: | (3E)-3-[(2,4-DIMETHOXYPHENYL)METHYLIDENE]-3,4,5,6-TETRAHYDRO-2,3'-BIPYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants for Interaction of Partial Agonists with Acetylcholine Binding Protein and Neuronal Alpha7 Nicotinic Acetylcholine Receptor.
Embo J., 28, 2009
|
|
2X2T
 
 | | CRYSTAL STRUCTURE OF SCLEROTINIA SCLEROTIORUM AGGLUTININ SSA in complex with Gal-beta1,3-Galnac | | Descriptor: | AGGLUTININ, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
2X34
 
 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | CELLULOSE-BINDING PROTEIN, X158, Ubiquinone-8 | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2X65
 
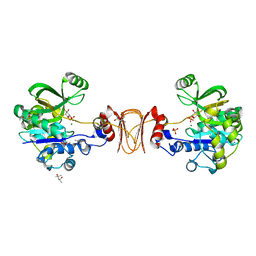 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with mannose-1-phosphate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, ... | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
2WN9
 
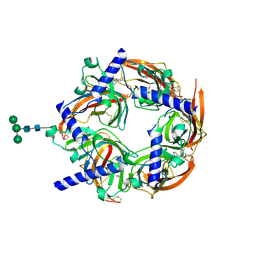 | | Crystal structure of Aplysia ACHBP in complex with 4-0H-DMXBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-5,6-DIHYDRO-2,3'-BIPYRIDIN-3(4H)-YLIDENEMETHYL]-3-METHOXYPHENOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
Embo J., 28, 2009
|
|
2X5S
 
 | |
2X2S
 
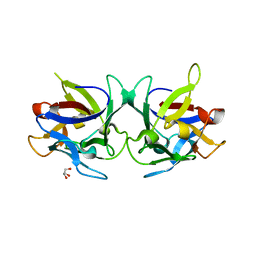 | | Crystal structure of Sclerotinia sclerotiorum agglutinin SSA | | Descriptor: | AGGLUTININ, GLYCEROL | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Rouge, P, Van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2010-01-15 | | Release date: | 2010-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Galnac/Gal-Specific Agglutinin from the Phytopathogenic Ascomycete Sclerotinia Sclerotiorum Reveals Novel Adaptation of a Beta-Trefoil Domain
J.Mol.Biol., 400, 2010
|
|
2WNL
 
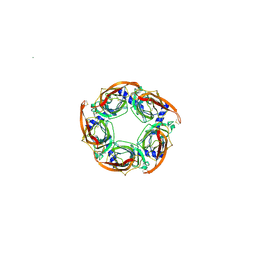 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH ANABASEINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4,5,6-tetrahydro-2,3'-bipyridine, 5-amino-1-pyridin-3-ylpentan-1-one, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
2WNC
 
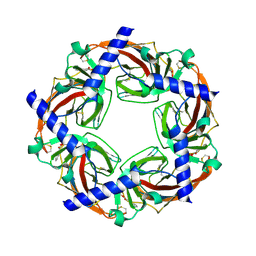 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
2X32
 
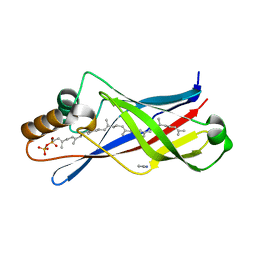 | | Structure of a polyisoprenoid binding domain from Saccharophagus degradans implicated in plant cell wall breakdown | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, CELLULOSE-BINDING PROTEIN, IMIDAZOLE | | Authors: | Vincent, F, Dal Molin, D, Weiner, R.M, Bourne, Y, Henrissat, B. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Polyisoprenoid Binding Domain from Saccharophagus Degradans Implicated in Plant Cell Wall Breakdown
FEBS Lett., 584, 2010
|
|
2X5Z
 
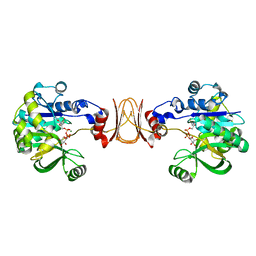 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GDP-mannose. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
2X60
 
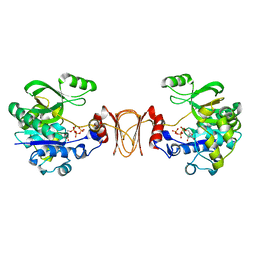 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANNOSE-1-PHOSPHATE GUANYLYLTRANSFERASE | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-12 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
2XB6
 
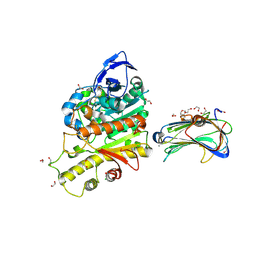 | | Revisited crystal structure of Neurexin1beta-Neuroligin4 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Leone, P, Comoletti, D, Ferracci, G, Conrod, S, Garcia, S.U, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Embo J., 29, 2010
|
|
2XBZ
 
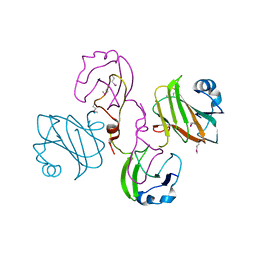 | | Multiple oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand-binding site | | Descriptor: | RETS-HYBRID SENSOR KINASE | | Authors: | Vincent, F, Round, A, Reynaud, A, Bordi, C, Filloux, A, Bourne, Y. | | Deposit date: | 2010-04-15 | | Release date: | 2010-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Distinct Oligomeric Forms of the Pseudomonas Aeruginosa Rets Sensor Domain Modulate Accessibility to the Ligand Binding Site.
Environ.Microbiol., 12, 2010
|
|
5MU5
 
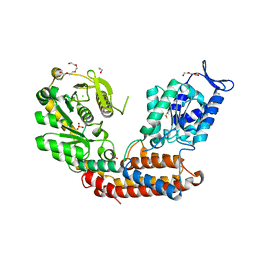 | | Structure of MAf glycosyltransferase from Magnetospirillum magneticum AMB-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Murat, D, Vincentelli, R, Wu, L.F, Guerardel, Y, Alberto, F. | | Deposit date: | 2017-01-12 | | Release date: | 2017-11-15 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylate and move! The glycosyltransferase Maf is involved in bacterial flagella formation.
Environ. Microbiol., 20, 2018
|
|
7QFU
 
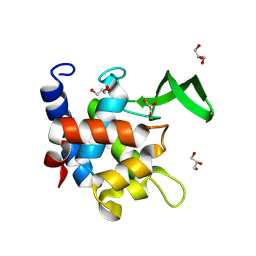 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
1AMY
 
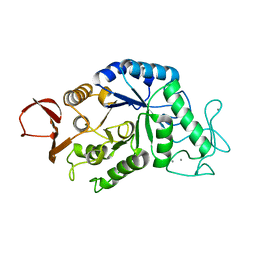 | |
7ZM1
 
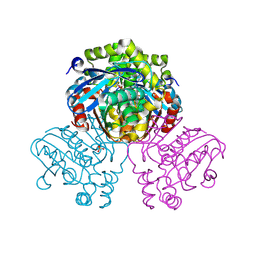 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC7b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(~{E},3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]tridec-11-enyl]phosphinous acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZM3
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC17 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, hexadecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
