1F21
 
 | |
1FNN
 
 | | CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6, MAGNESIUM ION | | Authors: | Liu, J, Smith, C.L, DeRyckere, D, DeAngelis, K, Martin, G.S, Berger, J.M. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Mol.Cell, 6, 2000
|
|
1Z5B
 
 | |
1Z59
 
 | | Topoisomerase VI-B, ADP-bound monomer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1Z5A
 
 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1Z5C
 
 | | Topoisomerase VI-B, ADP Pi bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1ZUN
 
 | | Crystal Structure of a GTP-Regulated ATP Sulfurylase Heterodimer from Pseudomonas syringae | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Mougous, J.D, Lee, D.H, Hubbard, S.C, Schelle, M.W, Vocadlo, D.J, Berger, J.M, Bertozzi, C.R. | | Deposit date: | 2005-05-31 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for g protein control of the prokaryotic ATP sulfurylase.
Mol.Cell, 21, 2006
|
|
1ZVU
 
 | |
1ZVT
 
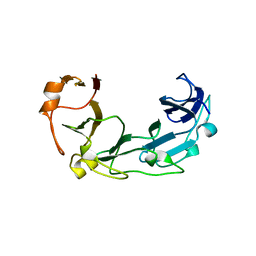 | |
2A8V
 
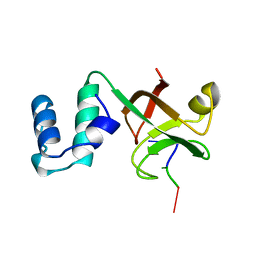 | | RHO TRANSCRIPTION TERMINATION FACTOR/RNA COMPLEX | | Descriptor: | 5'-R(P*CP*CP*C)-3', 5'-R(P*CP*CP*CP*CP*CP*C)-3', RNA BINDING DOMAIN OF RHO TRANSCRIPTION TERMINATION FACTOR | | Authors: | Bogden, C.E, Fass, D, Bergman, N, Nichols, M.D, Berger, J.M. | | Deposit date: | 1998-11-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for terminator recognition by the Rho transcription termination factor.
Mol.Cell, 3, 1999
|
|
2A11
 
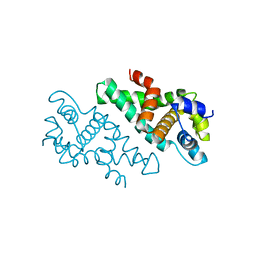 | |
2AU3
 
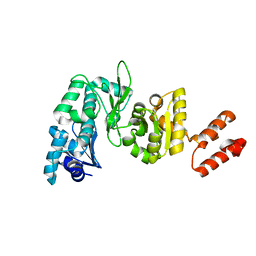 | | Crystal Structure of the Aquifex aeolicus primase (Zinc Binding and RNA Polymerase Domains) | | Descriptor: | DNA primase, ZINC ION | | Authors: | Corn, J.E, Pease, P.J, Hura, G.L, Berger, J.M. | | Deposit date: | 2005-08-26 | | Release date: | 2005-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crosstalk between primase subunits can act to regulate primer synthesis in trans.
Mol.Cell, 20, 2005
|
|
3RRN
 
 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3GFP
 
 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HVD
 
 | | The Protective Antigen Component of Anthrax Toxin Forms Functional Octameric Complexes | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Kintzer, A.F, Dong, K.C, Berger, J.M, Krantz, B.A. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The protective antigen component of anthrax toxin forms functional octameric complexes.
J.Mol.Biol., 392, 2009
|
|
3ICE
 
 | | Rho transcription termination factor bound to RNA and ADP-BeF3 | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Thomsen, N.D, Berger, J.M. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Running in reverse: the structural basis for translocation polarity in hexameric helicases.
Cell(Cambridge,Mass.), 139, 2009
|
|
3TO1
 
 | | Two surfaces on Rtt106 mediate histone binding and chaperone activity | | Descriptor: | Histone chaperone RTT106 | | Authors: | Zunder, R.M, Antczak, A.J, Berger, J.M, Rine, J. | | Deposit date: | 2011-09-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IBP
 
 | | The Crystal Structure of the Dimerization Domain of Escherichia coli Structural Maintenance of Chromosomes Protein MukB | | Descriptor: | AMMONIUM ION, Chromosome partition protein mukB | | Authors: | Li, Y, Schoeffler, A.J, Berger, J.M, Oakley, M.G. | | Deposit date: | 2009-07-16 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB.
J.Mol.Biol., 395, 2010
|
|
3ILW
 
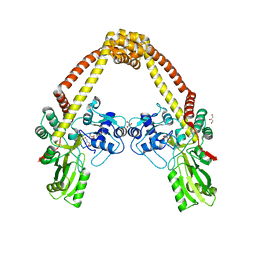 | | Structure of DNA gyrase subunit A N-terminal domain | | Descriptor: | DNA gyrase subunit A, GLYCEROL | | Authors: | Tretter, E.M, Schoeffler, A.J, Weisfield, S.R, Berger, J.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-07 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Crystal structure of the DNA gyrase GyrA N-terminal domain from Mycobacterium tuberculosis.
Proteins, 78, 2010
|
|
3K60
 
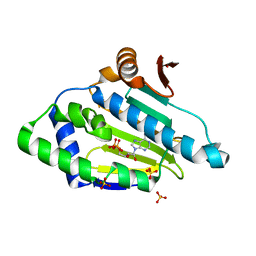 | |
3KDE
 
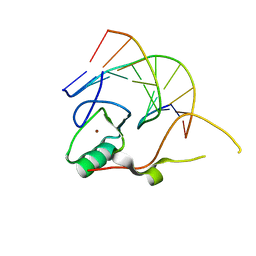 | | Crystal structure of the THAP domain from D. melanogaster P-element transposase in complex with its natural DNA binding site | | Descriptor: | 5'-D(*(BRU)P*CP*CP*AP*CP*TP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*AP*GP*(BRU)P*GP*GP*A)-3', Transposable element P transposase, ... | | Authors: | Sabogal, A, Lyubimov, A.Y, Berger, J.M, Rio, D.C. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KML
 
 | |
3L4J
 
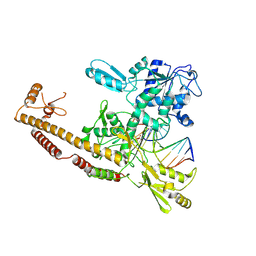 | | Topoisomerase II-DNA cleavage complex, apo | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
3L4K
 
 | | Topoisomerase II-DNA cleavage complex, metal-bound | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
