7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AP6
 
 | | Structure of SARS-CoV-2 Main Protease bound to MUT056399. | | Descriptor: | 3C-like proteinase, 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWS
 
 | | Structure of SARS-CoV-2 Main Protease bound to TH-302. | | Descriptor: | 3C-like proteinase, 5-[[(2-bromoethylamino)-(ethylamino)phosphoryl]oxymethyl]-1-methyl-~{N},~{N}-bis(oxidanyl)imidazol-2-amine, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
7N37
 
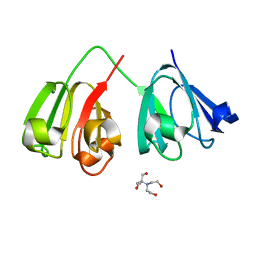 | | Crystal structure of 3-site deamidated variant of human gamma(S)-crystallin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma-crystallin S, MAGNESIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Martin, R.W. | | Deposit date: | 2021-05-31 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Deamidation of the human eye lens protein gamma S-crystallin accelerates oxidative aging.
Structure, 30, 2022
|
|
7N39
 
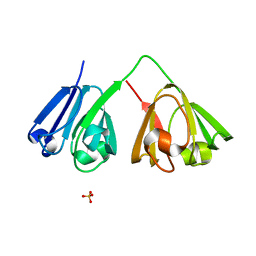 | |
7N38
 
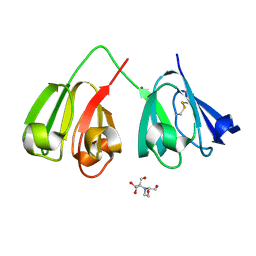 | | Crystal structure of 5-site deamidated variant of human gamma(S)-crystallin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma-crystallin S, MAGNESIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Martin, R.W. | | Deposit date: | 2021-05-31 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Deamidation of the human eye lens protein gamma S-crystallin accelerates oxidative aging.
Structure, 30, 2022
|
|
7N3B
 
 | |
7N36
 
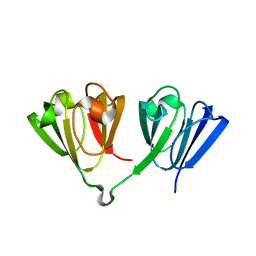 | |
7N3A
 
 | |
7NJJ
 
 | | Proteinase K grown inside HARE serial crystallography chip | | Descriptor: | NITRATE ION, Proteinase K | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJE
 
 | | gamma(S)-crystallin 9-site deamidation mutant grown inside HARE serial crystallography chip | | Descriptor: | Gamma-crystallin S | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJH
 
 | | HEX1 (in cellulo) grown inside HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJF
 
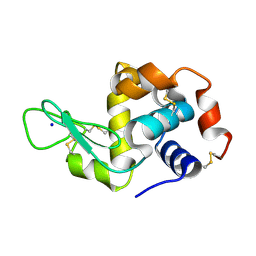 | | Hen egg white lysozyme (HEWL) grown inside HARE serial crystallography chip | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJG
 
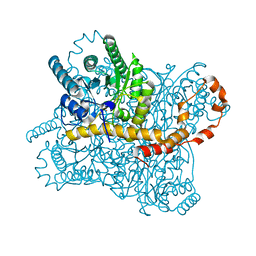 | | Xylose isomerase grown inside HARE serial crystallography chip | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJI
 
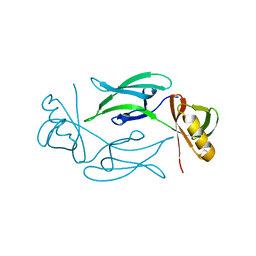 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NKF
 
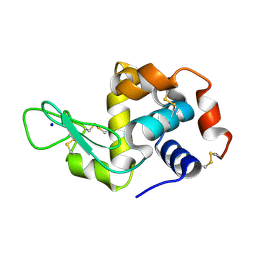 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6BEN
 
 | | Solution structure of de novo macrocycle design8.2 | | Descriptor: | (DAR)Q(DPR)(DGN)R(DGL)PQ | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, B. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BE9
 
 | | Solution structure of de novo macrocycle design7.1 | | Descriptor: | T(DLY)NDT(DSG)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, B. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
