1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1BUJ
 
 | |
5LQV
 
 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
1NOR
 
 | | TWO-DIMENSIONAL 1H-NMR STUDY OF THE SPATIAL STRUCTURE OF NEUROTOXIN II FROM NAJA OXIANA | | Descriptor: | NEUROTOXIN II | | Authors: | Golovanov, A.P, Utkin, Y.N, Lomize, A.L, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 1993-04-05 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H-NMR study of the spatial structure of neurotoxin II from Naja naja oxiana.
Eur.J.Biochem., 213, 1993
|
|
1RQU
 
 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQS
 
 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RL5
 
 | | NMR structure with tightly bound water molecule of cytotoxin I from Naja oxiana in aqueous solution (major form) | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2003-11-25 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins.
Biochem.J., 387, 2005
|
|
1SCO
 
 | | SCORPION TOXIN (OSK1 TOXIN) WITH HIGH AFFINITY FOR SMALL CONDUCTANCE CA(2+)-ACTIVATED K+ CHANNEL IN NEUROBLASTOMA-X-GLUOMA NG 108-15 HYBRID CELLS, NMR, 30 STRUCTURES | | Descriptor: | SCORPION TOXIN OSK1 | | Authors: | Jaravine, V.A, Nolde, D.E, Pluzhnikov, K.A, Grishin, E.V, Arseniev, A.S. | | Deposit date: | 1996-04-01 | | Release date: | 1997-01-27 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of toxin OSK1 from Orthochirus scrobiculosus scorpion venom.
Biochemistry, 36, 1997
|
|
1RQT
 
 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQV
 
 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
8AR3
 
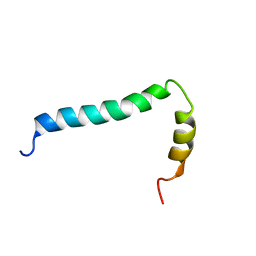 | | Solution structure of TLR9 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 9 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
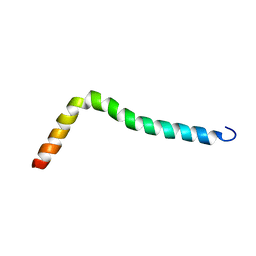 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR1
 
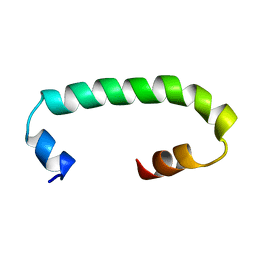 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR2
 
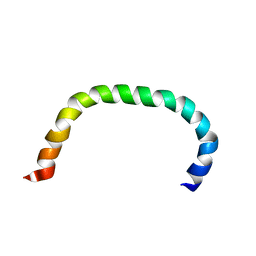 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
2G9P
 
 | | NMR structure of a novel antimicrobial peptide, latarcin 2a, from spider (Lachesana tarabaevi) venom | | Descriptor: | antimicrobial peptide Latarcin 2a | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2006-03-07 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Spatial structure and activity mechanism of a novel spider antimicrobial peptide.
Biochemistry, 45, 2006
|
|
2J5D
 
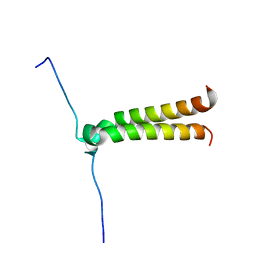 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
2FR9
 
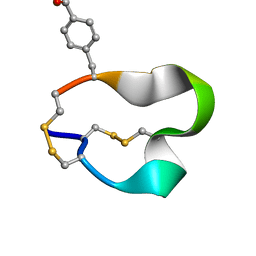 | | NMR structure of the alpha-conotoxin GI (SER12)-benzoylphenylalanine derivative | | Descriptor: | Alpha-conotoxin GI | | Authors: | Pashkov, V.S, Maslennikov, I.V, Kasheverov, I.E, Zhmak, M.N, Utkin, Y.N, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Alpha-Conotoxin GI benzoylphenylalanine derivatives. (1)H-NMR structures and photoaffinity labeling of the Torpedo californica nicotinic acetylcholine receptor.
Febs J., 273, 2006
|
|
2FRB
 
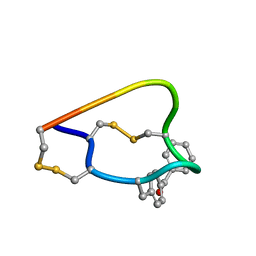 | | NMR structure of the alpha-conotoxin GI (ASN4)-benzoylphenylalanine derivative | | Descriptor: | Alpha-conotoxin GIA | | Authors: | Pashkov, V.S, Maslennikov, I, Kasheverov, I.V, Zhmak, M.N, Utkin, Y.N, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Alpha-Conotoxin GI benzoylphenylalanine derivatives. (1)H-NMR structures and photoaffinity labeling of the Torpedo californica nicotinic acetylcholine receptor.
Febs J., 273, 2006
|
|
2JWM
 
 | |
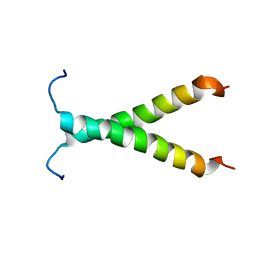
2K1L
 
 | |
2JWA
 
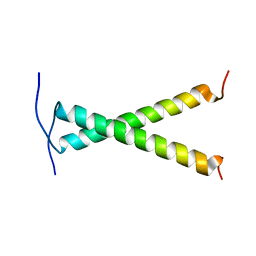 | |
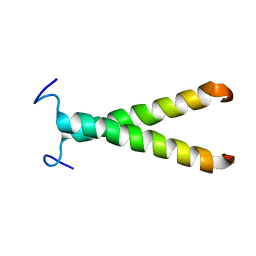
2K1K
 
 | |
2JNI
 
 | | Spatial structure of antimicrobial peptide arenicin-2 in aqueous solution | | Descriptor: | Arenicin-2 | | Authors: | Ovchinnikova, T.V, Shenkarev, Z.O, Nadezhdin, K.D, Balandin, S.V, Zhmak, M.N, Kudelina, I.A, Finkina, E.I, Kokryakov, V.N, Arseniev, A.S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Recombinant expression, synthesis, purification, and solution structure of arenicin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
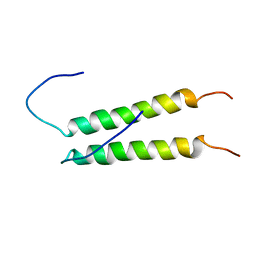
2K9Y
 
 | |
