7K2F
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GAEETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GAEETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2R
 
 | | Kelch domain of human KEAP1 bound to Nrf2-based cyclic peptide, c[LhA-DEETGE] | | Descriptor: | B3A-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2I
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[GAPETGE] | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 cyclic peptide,c[GAPETGE] | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2D
 
 | | Kelch domain of human KEAP1 bound to Nrf2 linear peptide, Ac-GDEETGE-NH2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 linear peptide, Ace-GDEETGE-NH2 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K2Q
 
 | | Kelch domain of human KEAP1 bound to Nrf2 cyclic peptide, c[Ahx-DPETGE] | | Descriptor: | ACA-ASP-PRO-GLU-THR-GLY-GLU, Kelch-like ECH-associated protein 1 | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
7K29
 
 | | Kelch domain of human KEAP1 bound to Nrf2 peptide, LDEETGEAL | | Descriptor: | ACE-LEU-ASP-GLU-GLU-THR-GLY-GLU-ALA-LEU-NH2, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Muellers, S.N, Allen, K.N. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recapitulating the Binding Affinity of Nrf2 for KEAP1 in a Cyclic Heptapeptide, Guided by NMR, X-ray Crystallography, and Machine Learning.
J.Am.Chem.Soc., 143, 2021
|
|
2IOH
 
 | | Crystal structure of phosphonoacetaldehyde hydrolase with a K53R mutation | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphonoacetaldehyde hydrolase | | Authors: | Allen, K.A, Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Peisach, E. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage.
Bioorg.Chem., 34, 2006
|
|
2IOF
 
 | | Crystal structure of phosphonoacetaldehyde hydrolase with sodium borohydride-reduced substrate intermediate | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Phosphonoacetaldehyde hydrolase | | Authors: | Allen, K.A, Lahiri, S.D, Zhang, G, Dunaway-Mariano, D. | | Deposit date: | 2006-10-10 | | Release date: | 2007-07-17 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage.
Bioorg.Chem., 34, 2006
|
|
4OLG
 
 | | Crystal structure of AmpC beta-lactamase in complex with covalently bound N-formyl 7-aminocephalosporanic acid | | Descriptor: | (2R,5Z)-5-[(acetyloxy)methylidene]-2-[(1R)-1-(formylamino)-2-oxoethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Shoichet, B.K, Barelier, S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-05-28 | | Last modified: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Substrate deconstruction and the nonadditivity of enzyme recognition.
J.Am.Chem.Soc., 136, 2014
|
|
4OKP
 
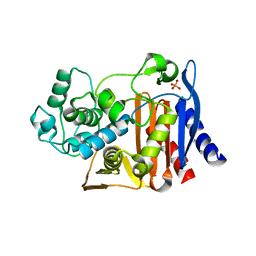 | |
4OLD
 
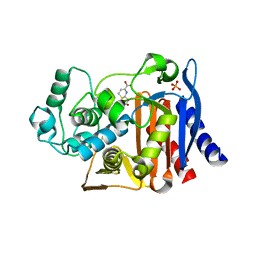 | | Crystal structure of AmpC beta-lactamase in complex with the product form of (6R,7R)-7-amino-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid | | Descriptor: | (2R)-2-[(R)-amino(carboxy)methyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Shoichet, B.K, Barelier, S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-05-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substrate deconstruction and the nonadditivity of enzyme recognition.
J.Am.Chem.Soc., 136, 2014
|
|
3MMZ
 
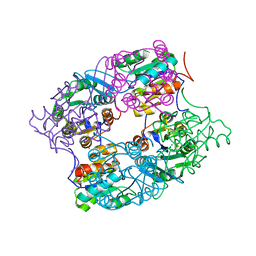 | | CRYSTAL STRUCTURE OF putative HAD family hydrolase from Streptomyces avermitilis MA-4680 | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative HAD family hydrolase | | Authors: | Malashkevich, V.N, Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3MN1
 
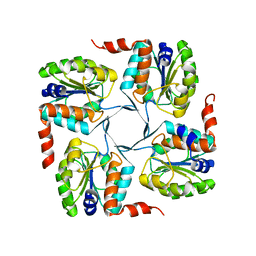 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3N07
 
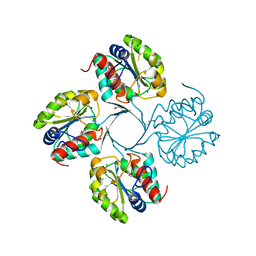 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
5TJR
 
 | |
5E0O
 
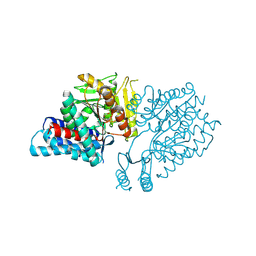 | |
3QU9
 
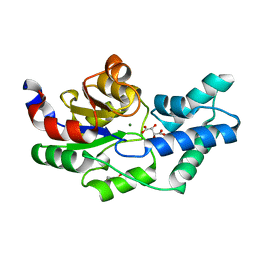 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with magnesium and tartrate | | Descriptor: | CHLORIDE ION, GLYCEROL, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QXG
 
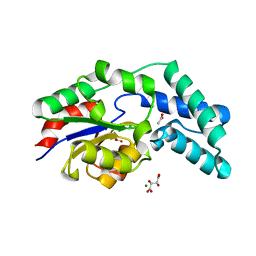 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron complexed with calcium, a closed cap conformation | | Descriptor: | ACETATE ION, CALCIUM ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-01 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU4
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13ala mutant | | Descriptor: | ACETATE ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUQ
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, an open cap conformation | | Descriptor: | CHLORIDE ION, FORMIC ACID, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-24 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QUB
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, glu47ala mutant complexed with sulfate | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QX7
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron complexed with phosphate, a closed cap conformation | | Descriptor: | INORGANIC PYROPHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-01 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU7
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp13asn mutant complexed with calcium and phosphate | | Descriptor: | ACETATE ION, CALCIUM ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU5
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, asp11asn mutant | | Descriptor: | CHLORIDE ION, INORGANIC PYROPHOSPHATASE | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
3QU2
 
 | | Crystal structure of pyrophosphatase from bacteroides thetaiotaomicron, a closed cap conformation | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Huang, H, Toro, R, Gerlt, J.A, Burley, S.K, Dunaway-Mariano, D, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), Enzyme Function Initiative (EFI) | | Deposit date: | 2011-02-23 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Divergence of Structure and Function in the Haloacid Dehalogenase Enzyme Superfamily: Bacteroides thetaiotaomicron BT2127 Is an Inorganic Pyrophosphatase.
Biochemistry, 50, 2011
|
|
