1H8U
 
 | | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A: An Atypical Lectin with a Paradigm Shift in Specificity | | Descriptor: | EOSINOPHIL GRANULE MAJOR BASIC PROTEIN 1, GLYCEROL, SULFATE ION | | Authors: | Swaminathan, G.J, Weaver, A.J, Loegering, D.A, Checkel, J.L, Leonidas, D.D, Gleich, G.J, Acharya, K.R. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Eosinophil Major Basic Protein at 1.8A. An Atypical Lectin with a Paradigm Shift in Specificity
J.Biol.Chem., 276, 2001
|
|
1HA5
 
 | | Structural features of a zinc-binding site in the superantigen streptococcal pyrogenic exotoxin A (SpeA1): implications for MHC class II recognition. | | Descriptor: | STREPTOCOCCAL PYOGENIC EXOTOXIN A1, ZINC ION | | Authors: | Baker, M.D, Gutman, D.M, Papageorgiou, A.C, Collins, C.M, Acharya, K.R. | | Deposit date: | 2001-03-28 | | Release date: | 2002-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Features of a Zinc Binding Site in the Superantigen Strepococcal Pyrogenic Exotoxin a (Spea1): Implications for Mhc Class II Recognition.
Protein Sci., 10, 2001
|
|
1H1H
 
 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | Deposit date: | 2002-07-15 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
2R3Z
 
 | | Crystal structure of mouse IP-10 | | Descriptor: | Small-inducible cytokine B10 | | Authors: | Jabeen, T, Leonard, P, Jamaluddin, H, Acharya, K.R. | | Deposit date: | 2007-08-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of mouse IP-10, a chemokine
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2SKE
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHITE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHITE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2SKC
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH FLUOROPHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | FLUORO-PHOSPHITE ION, INOSINIC ACID, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2SKD
 
 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHATE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHATE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
2VFZ
 
 | | CRYSTAL STRUCTURE OF ALPHA-1,3 GALACTOSYLTRANSFERASE (R365K) IN COMPLEX WITH UDP-2F-GALACTOSE | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2007-11-06 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis
J.Mol.Biol., 369, 2007
|
|
2VS3
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2VS4
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2VS5
 
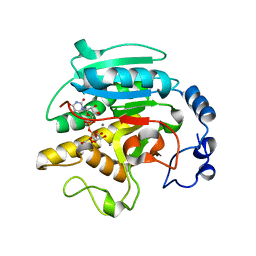 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
2W5L
 
 | | RNASE A-NADP COMPLEX | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2VWE
 
 | | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralizing Antibody Fab Fragment | | Descriptor: | ANTI-VEGF-B MONOCLONAL ANTIBODY, VASCULAR ENDOTHELIAL GROWTH FACTOR B | | Authors: | Leonard, P, Scotney, P.D, Jabeen, T, Iyer, S, Fabri, L.J, Nash, A.D, Acharya, K.R. | | Deposit date: | 2008-06-23 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralising Antibody Fab Fragment.
J.Mol.Biol., 384, 2008
|
|
2VXL
 
 | | Screening a Limited Structure-based Library Identifies UDP-GalNAc- Specific Mutants of alpha-1,3 Galactosyltransferase | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Acharya, K.R, Brew, K. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Screening a Limited Structure-Based Library Identifies Udp-Galnac-Specific Mutants of {Alpha}-1,3-Galactosyltransferase.
Glycobiology, 18, 2008
|
|
2W5G
 
 | | RNASE A-5'-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate- Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2WD2
 
 | | A chimeric microtubule disruptor with efficacy on a taxane resistant cell line | | Descriptor: | 7-methoxy-2-(3-methoxybenzyl)-1,2,3,4-tetrahydroisoquinolin-6-yl sulfamate, CARBONIC ANHYDRASE 2, FORMIC ACID, ... | | Authors: | Leese, M.P, Jourdan, F.L, Kimberley, M.R, Cozier, G.E, Regis-Lydi, S, Foster, P.A, Newman, S.P, Thiyagarajan, N, Acharya, K.R, Ferrandis, E, Purohit, A, Reed, M.J, Potter, B.V.L. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chimeric Microtubule Disruptors.
Chem.Commun.(Camb.), 46, 2010
|
|
2WGZ
 
 | | Crystal structure of alpha-1,3 galactosyltransferase (alpha3GT) in a complex with p-nitrophenyl-beta-galactoside (pNP-beta-Gal) | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jamaluddin, H, Tumbale, P, Ferns, T.A, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2009-04-28 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Alpha-1,3 Galactosyltransferase (Alpha3Gt) in a Complex with P-Nitrophenyl-Beta-Galactoside (Pnpbetagal).
Biochem.Biophys.Res.Commun., 385, 2009
|
|
2W2D
 
 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2VQ9
 
 | | RNASE ZF-3E | | Descriptor: | CHLORIDE ION, RNASE 1 | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications
J.Mol.Biol., 380, 2008
|
|
2VQ8
 
 | | RNASE ZF-1A | | Descriptor: | CHLORIDE ION, RNASE ZF-1A | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications.
J.Mol.Biol., 380, 2008
|
|
2W5K
 
 | | RNASE A-NADPH COMPLEX | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2W5M
 
 | | RNASE A-PYROPHOSPHATE ION COMPLEX | | Descriptor: | PYROPHOSPHATE 2-, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2VXM
 
 | | Screening a Limited Structure-based Library Identifies UDP-GalNAc- Specific Mutants of alpha-1,3 Galactosyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Acharya, K.R, Brew, K. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Screening a Limited Structure-Based Library Identifies Udp-Galnac-Specific Mutants of {Alpha}-1,3-Galactosyltransferase.
Glycobiology, 18, 2008
|
|
2W5I
 
 | | RNASE A-AP3A COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2WD3
 
 | | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors based on a Biphenyl Template | | Descriptor: | 3-CHLORO-2'-CYANO-5'-(1H-1,2,4-TRIAZOL-1-YLMETHYL)BIPHENYL-4-YL SULFAMATE, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woo, L.W.L, Jackson, T, Putey, A, Cozier, G, Leonard, P, Acharya, K.R, Chander, S.K, Purohit, A, Reed, M.J, Potter, B.V.L. | | Deposit date: | 2009-03-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly Potent First Examples of Dual Aromatase-Steroid Sulfatase Inhibitors Based on a Biphenyl Template.
J.Med.Chem., 53, 2010
|
|
