7F59
 
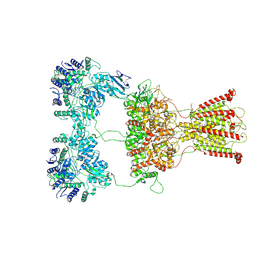 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F56
 
 | | DNQX-bound GluK2-1xNeto2 complex, with asymmetric LBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F57
 
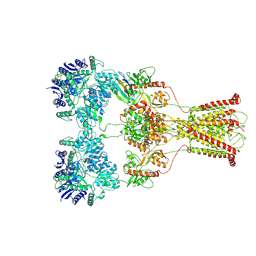 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5B
 
 | | LBD-TMD focused reconstruction of DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5A
 
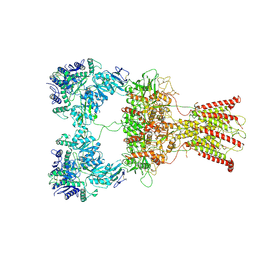 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7XAF
 
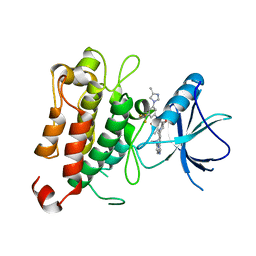 | | The crystal structure of TrkA kinase in complex with 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo- tetradecaphan-2-yne-45-carboxamide | | Descriptor: | 4^6,14-dimethyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-10-oxo-5-oxa-11,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclo-tetradecaphan-2-yne-45-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-03-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001182 Å) | | Cite: | Discovery of the First Highly Selective and Broadly Effective Macrocycle-Based Type II TRK Inhibitors that Overcome Clinically Acquired Resistance.
J.Med.Chem., 65, 2022
|
|
8HJD
 
 | |
8HJC
 
 | |
8HIJ
 
 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
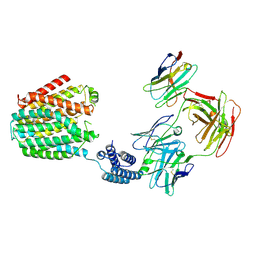 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
7YLX
 
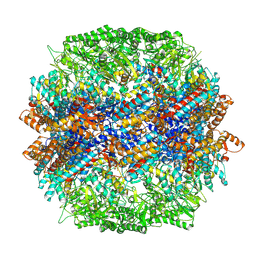 | | yeast TRiC-plp2-actin complex at S4 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLY
 
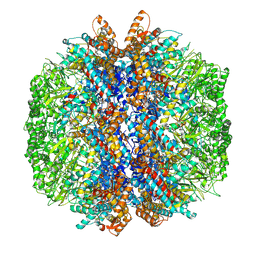 | | yeast TRiC-plp2 complex at S5 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLV
 
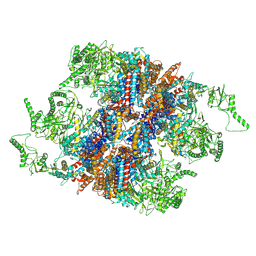 | | yeast TRiC-plp2-substrate complex at S2 ATP binding state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
6ATH
 
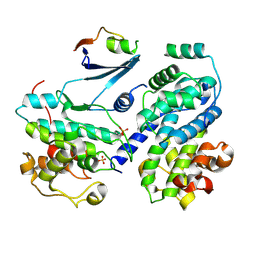 | | Cdk2/cyclin A/p27-KID-deltaC | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation.
Nat Commun, 10, 2019
|
|
7YLU
 
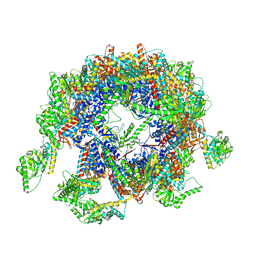 | | yeast TRiC-plp2-substrate complex at S1 TRiC-NPP state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLW
 
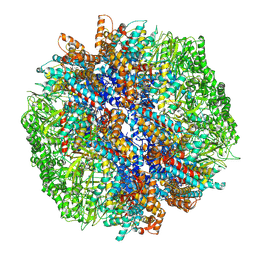 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
6BTM
 
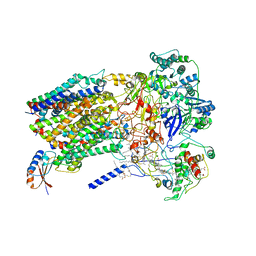 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
6APA
 
 | |
7XML
 
 | | Cryo-EM structure of PEIP-Bs_enolase complex | | Descriptor: | Enolase, MAGNESIUM ION, Putative gene 60 protein | | Authors: | Li, S, Zhang, K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage protein PEIP is a potent Bacillus subtilis enolase inhibitor.
Cell Rep, 40, 2022
|
|
4HIQ
 
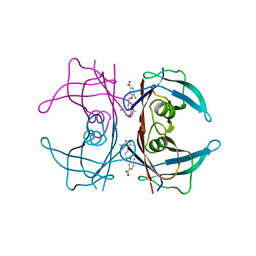 | | The Structure of V122I Mutant Transthyretin in Complex with AG10 | | Descriptor: | 3-[3-(3,5-dimethyl-1H-pyrazol-4-yl)propoxy]-4-fluorobenzoic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HIS
 
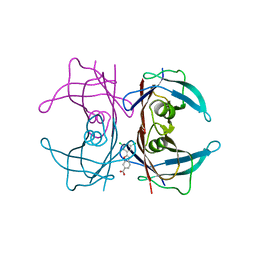 | | The Structure of V122I Mutant Transthyretin in Complex with Tafamidis | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Connelly, S, Alhamadsheh, M, Graef, I, Wilson, I.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7WJ6
 
 | |
7WJ7
 
 | |
4IWY
 
 | | SeMet-substituted RimK structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
