7QPJ
 
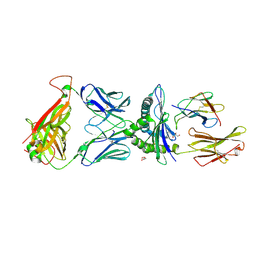 | | Crystal structure of engineered TCR (756) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
6CPW
 
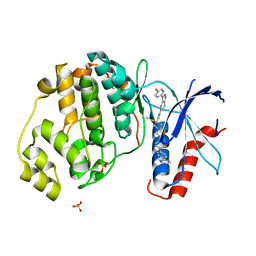 | | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology | | Descriptor: | (3S)-N-[3-(4-fluorophenyl)-1H-indazol-5-yl]-3-(methylsulfanyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Hruza, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6B5P
 
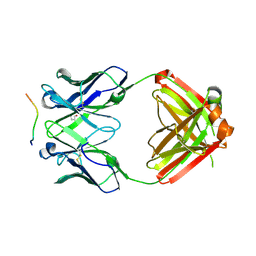 | | Structure of PfCSP peptide 20 with human antibody CIS42 | | Descriptor: | CIS42 Fab Heavy chain, CIS42 Fab Light chain, pfCSP peptide 20: ASN-PRO-ASP-PRO-ASN-ALA-ASN-PRO-ASN-VAL-ASP | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6B5R
 
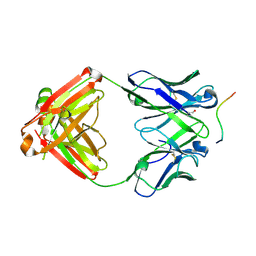 | | Structure of PfCSP peptide 21 with human antibody CIS42 | | Descriptor: | CIS42 Fab Heavy chain, CIS42 Fab Light chain, PfCSP peptide 21: ASN-PRO-ASP-PRO-ASN-ALA-ASN-PRO-ASN-VAL-ASP-PRO-ASN | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
5BUU
 
 | | Crystal structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM-321 at 2.07 A resolution | | Descriptor: | (3R)-7-chloro-2,3,4-trimethyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, 1,2-ETHANEDIOL, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Tapken, D, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors.
Acs Chem Neurosci, 7, 2016
|
|
5D9Q
 
 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 and scFv NIH45-46 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Ward, A.B, Wilson, I.A. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
4BL9
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLB
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8UH4
 
 | |
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BL8
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Karlstrom, M, Finta, C, Cherry, A.L, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9LPR
 
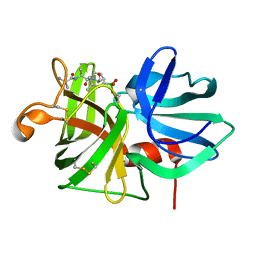 | |
1P10
 
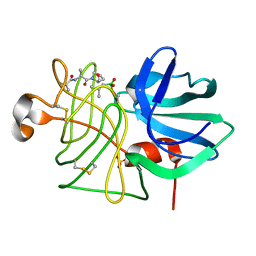 | |
1P03
 
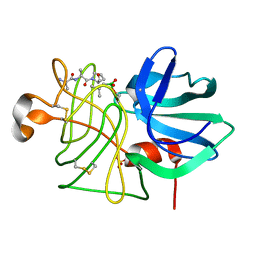 | |
1P09
 
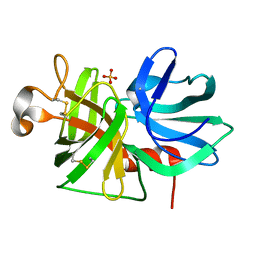 | |
1P04
 
 | |
1P05
 
 | |
1P06
 
 | |
1P02
 
 | |
1QBB
 
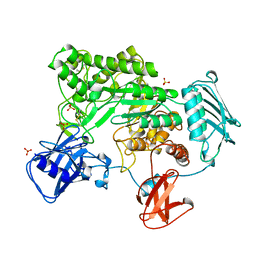 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBA
 
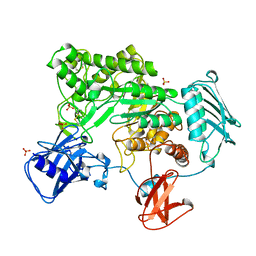 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
2KJ3
 
 | | High-resolution structure of the HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Van Melckebeke, H, Wasmer, C, Lange, A, AB, E, Loquet, A, Meier, B.H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of HET-s(218-289) Amyloid Fibrils by Solid-State NMR Spectroscopy
J.Am.Chem.Soc., 132, 2010
|
|
2I4P
 
 | |
