1KKM
 
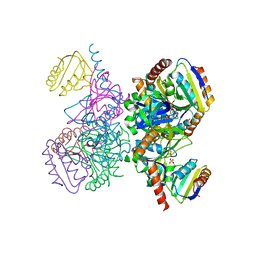 | | L.casei HprK/P in complex with B.subtilis P-Ser-HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHATE ION, ... | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1NVK
 
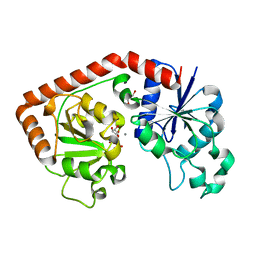 | | T4 phage BGT in complex with UDP and a Mn2+ ion at 1.8 A resolution | | Descriptor: | DNA beta-glucosyltransferase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lariviere, L, Kurzeck, J, Gueguen-Chaignon, V, Rueger, W, Morera, S. | | Deposit date: | 2003-02-04 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism
J.Mol.Biol., 330, 2003
|
|
1NZF
 
 | | T4 phage BGT-D100A mutant in complex with UDP-glucose: Form II | | Descriptor: | CHLORIDE ION, DNA beta-glycosyltransferase, GLYCEROL, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with
UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
1NZD
 
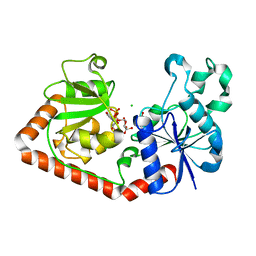 | | T4 phage BGT-D100A mutant in complex with UDP-glucose: Form I | | Descriptor: | CHLORIDE ION, DNA beta-glycosyltransferase, GLYCEROL, ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism
J.Mol.Biol., 330, 2003
|
|
1NDK
 
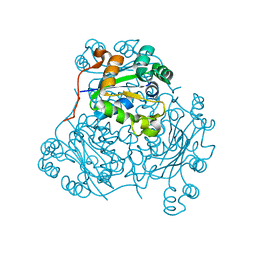 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
1M5R
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Base-flipping mechanism for the T4 phage beta-glucosyltransferase and
identification of a transition state analog
J.Mol.Biol., 324, 2002
|
|
1IXY
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', 5'-D(*GP*AP*TP*AP*CP*TP*3DRP*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Base-flipping Mechanism for the T4 Phage beta-Glucosyltransferase and Identification of a
Transition-state Analog
J.Mol.Biol., 324, 2002
|
|
1J39
 
 | | Crystal Structure of T4 phage BGT in complex with its UDP-glucose substrate | | Descriptor: | DNA beta-glucosyltransferase, GLYCEROL, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the T4 phage beta-glucosyltransferase and the D100A mutant in complex with UDP-glucose: glucose binding and identification of the catalytic base for a direct displacement mechanism.
J.Mol.Biol., 330, 2003
|
|
8CJ9
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CK6
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CKQ
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CKT
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CLW
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CM2
 
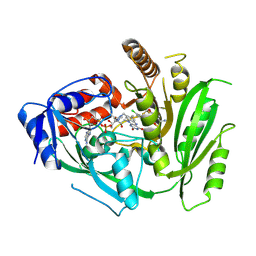 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[[3,5-dichloro-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
1LWX
 
 | | AZT DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1997-04-30 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of azido-thymidine diphosphate binding to nucleoside diphosphate kinase.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1B99
 
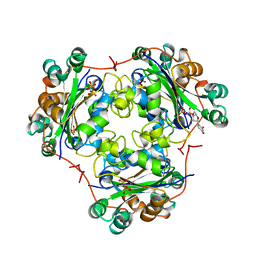 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1999-02-22 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
6D97
 
 | |
3BBC
 
 | |
1B4S
 
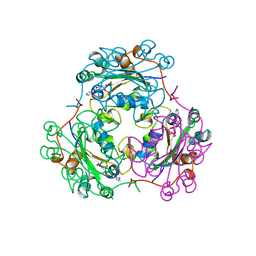 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE H122G MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE, ... | | Authors: | Meyer, P, Janin, J. | | Deposit date: | 1998-12-28 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase.
Biochemistry, 38, 1999
|
|
1BUX
 
 | | 3'-PHOSPHORYLATED NUCLEOTIDES BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Xu, Y, Schneider, B, Deville-Bonne, D, Veron, M, Janin, J. | | Deposit date: | 1998-09-07 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3'-Phosphorylated nucleotides are tight binding inhibitors of nucleoside diphosphate kinase activity.
J.Biol.Chem., 273, 1998
|
|
2FEP
 
 | | Structure of truncated CcpA in complex with P-Ser-HPr and Sulfate ions | | Descriptor: | Catabolite control protein A, Phosphocarrier protein HPr, SULFATE ION | | Authors: | Chaptal, V, Gueguen-Chaignon, V, Poncet, S, Lecampion, C, Meyer, P, Deutscher, J, Galinier, A, Nessler, S. | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of B. subtilis CcpA effector binding site.
Proteins, 64, 2006
|
|
2MJF
 
 | | Solution structure of the complex between the yeast Rsa1 and Hit1 proteins | | Descriptor: | Protein HIT1, Ribosome assembly 1 protein | | Authors: | Quinternet, M, Roth, B, Back, R, Jacquemin, C, Manival, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction.
Nucleic Acids Res., 42, 2014
|
|
2BEF
 
 | | CRYSTAL STRUCTURE OF NDP KINASE COMPLEXED WITH MG, ADP, AND BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, Y.W, Cherfils, J. | | Deposit date: | 1998-05-26 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
3BBF
 
 | | Crystal structure of the NM23-H2 transcription factor complex with GDP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1.
Mol.Cancer Ther., 8, 2009
|
|
3BBB
 
 | |
