2A3R
 
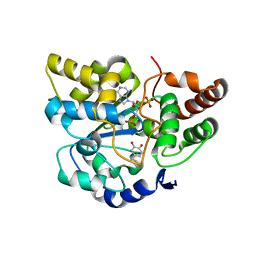 | | Crystal Structure of Human Sulfotransferase SULT1A3 in Complex with Dopamine and 3-Phosphoadenosine 5-Phosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, L-DOPAMINE, Monoamine-sulfating phenol sulfotransferase | | Authors: | Lu, J.H, Li, H.T, Liu, M.C, Zhang, J.P, Li, M, An, X.M, Chang, W.R. | | Deposit date: | 2005-06-26 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3'-phosphoadenosine 5'-phosphate
Biochem.Biophys.Res.Commun., 335, 2005
|
|
1FMX
 
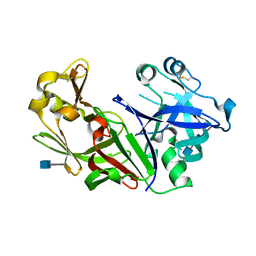 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1FMU
 
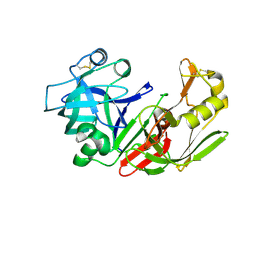 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1G0V
 
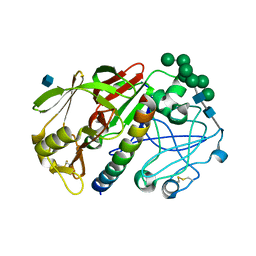 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
1GA1
 
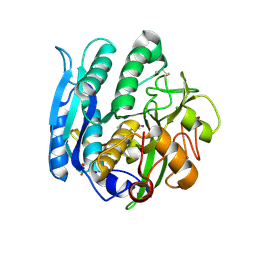 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF IODOTYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FRAGMENT OF IODOTYROSTATIN, ... | | Authors: | Dauter, Z, Li, M, Wlodawer, A. | | Deposit date: | 2000-11-29 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1B8Q
 
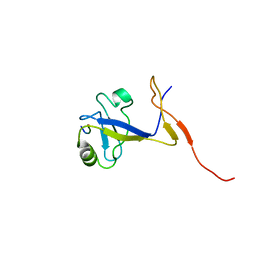 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
5FTE
 
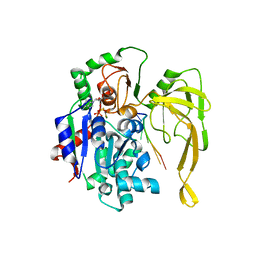 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTC
 
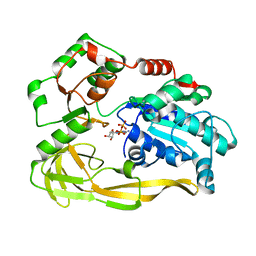 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTD
 
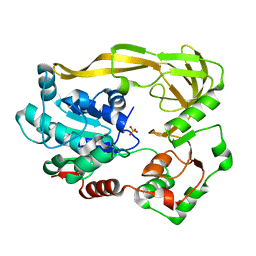 | | Crystal structure of Pif1 helicase from Bacteroides apo form | | Descriptor: | PHOSPHATE ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTF
 
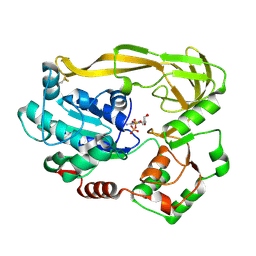 | | Crystal structure of Pif1 helicase from Bacteroides double mutant L95C-I339C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
1F95
 
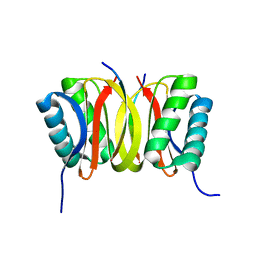 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND BIM PEPTIDE COMPLEX | | Descriptor: | BCL2-LIKE 11 (APOPTOSIS FACILITATOR), DYNEIN | | Authors: | Fan, J.-S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
1F96
 
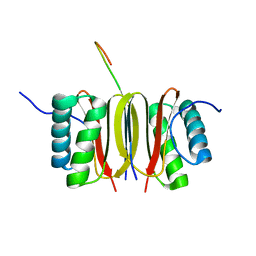 | | SOLUTION STRUCTURE OF DYNEIN LIGHT CHAIN 8 (DLC8) AND NNOS PEPTIDE COMPLEX | | Descriptor: | DYNEIN LIGHT CHAIN 8, PROTEIN (NNOS, NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Fan, J.S, Zhang, Q, Tochio, H, Li, M, Zhang, M. | | Deposit date: | 2000-07-07 | | Release date: | 2001-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain.
J.Mol.Biol., 306, 2001
|
|
5FTB
 
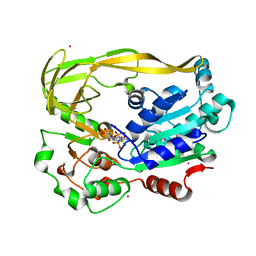 | | Crystal structure of Pif1 helicase from Bacteroides in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
1SN7
 
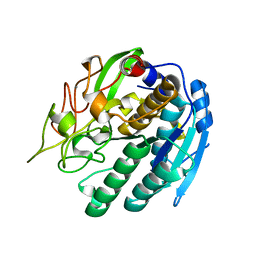 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
4R4X
 
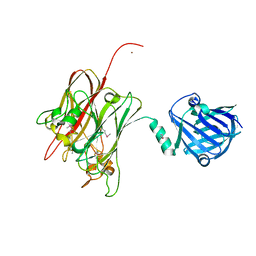 | | Structure of PNGF-II in C2 space group | | Descriptor: | PNGF-II, ZINC ION | | Authors: | Sun, G, Yu, X, Celimuge, Wang, L, Li, M, Gan, J, Qu, D, Ma, J, Chen, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and
Characterization of a Novel Prokaryotic Peptide: N-glycosidase from
Elizabethkingia meningoseptica
J.Biol.Chem., 2015
|
|
4RTH
 
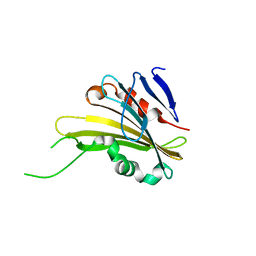 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RT4
 
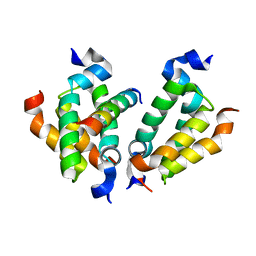 | | Crystal structure of Dpy30 complexed with Bre2 | | Descriptor: | Peptide from COMPASS component BRE2, Protein dpy-30 homolog | | Authors: | Zhang, H.M, Li, M, Chang, W.R. | | Deposit date: | 2014-11-12 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural implications of Dpy30 oligomerization for MLL/SET1 COMPASS H3K4 trimethylation
Protein Cell, 6, 2015
|
|
4RTI
 
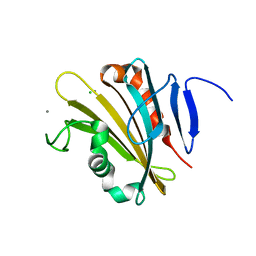 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
1P1E
 
 | |
4RTA
 
 | |
1P1D
 
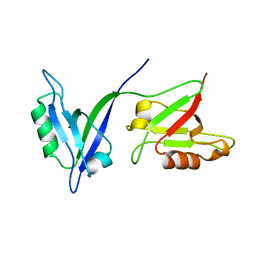 | |
2ZNX
 
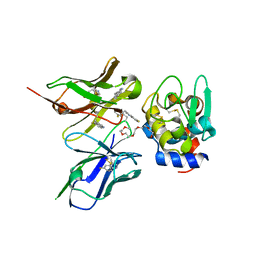 | | 5-Fluorotryptophan Incorporated ScFv10 Complexed to Hen Egg Lysozyme | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lysozyme C, ScFv | | Authors: | DeSantis, M.E, Acchione, M, Li, M, Walter, R.L, Wlodawer, A, Smith-Gill, S. | | Deposit date: | 2008-05-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics
Biochemistry, 51, 2012
|
|
4U4F
 
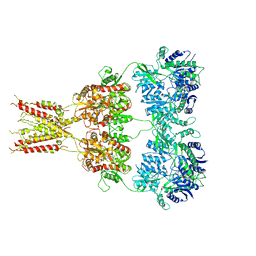 | | Structure of GluA2* in complex with partial agonist (S)-5-Nitrowillardiine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, ... | | Authors: | Yelshanskaya, M.V, Li, M, Sobolevsky, A.I. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.79 Å) | | Cite: | Structure of an agonist-bound ionotropic glutamate receptor.
Science, 345, 2014
|
|
8IHO
 
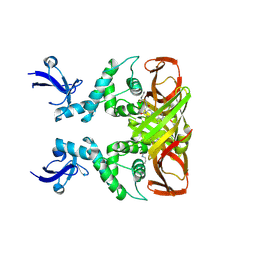 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
4U4G
 
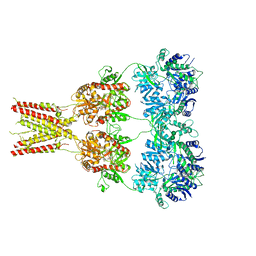 | | Structure of GluA2* in complex with competitive antagonist ZK 200775 | | Descriptor: | Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Yelshanskaya, M.V, Li, M, Sobolevsky, A.I. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.49 Å) | | Cite: | Structure of an agonist-bound ionotropic glutamate receptor.
Science, 345, 2014
|
|
