8J9K
 
 | | Structure of basal beta-arrestin2 | | Descriptor: | Beta-arrestin-2, Fab6 heavy chain, Fab6 light chain | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8Z
 
 | | Structure of beta-arrestin1 in complex with D6Rpp | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8V
 
 | | Structure of beta-arrestin2 in complex with D6Rpp (Local Refine) | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-2, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JA3
 
 | | Structure of beta-arrestin1 in complex with C3aRpp | | Descriptor: | Beta-arrestin-1, C3a anaphylatoxin chemotactic receptor, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
4QL1
 
 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
7OMT
 
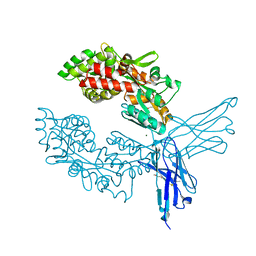 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
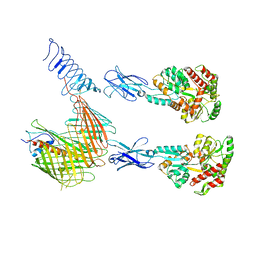 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
3V01
 
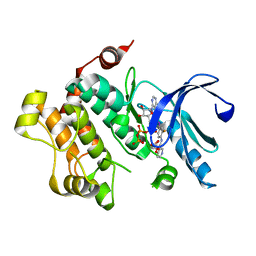 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
7AJQ
 
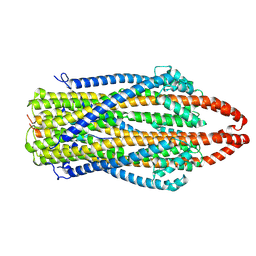 | | cryo-EM structure of ExbBD from Serratia Marcescens | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD | | Authors: | Biou, V, Adaixo, R, Coureux, P.D, Delepelaire, P, Chami, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
6NFT
 
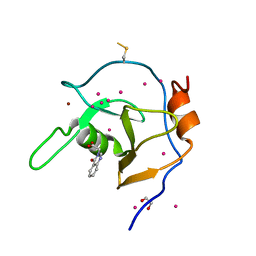 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (4-oxoquinazolin-3(4H)-yl)acetic acid | | Descriptor: | (4-oxoquinazolin-3(4H)-yl)acetic acid, 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Mann, M.K, Tempel, W, Bountra, C, Arrowmsmith, C.M, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-20 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
6YE4
 
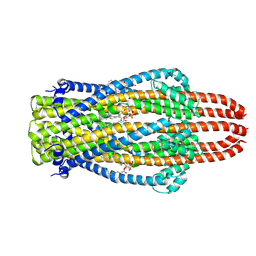 | | Structure of ExbB pentamer from Serratia marcescens by single particle cryo electron microscopy | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB | | Authors: | Biou, V, Delepelaire, P, Coureux, P.D, Chami, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
3V04
 
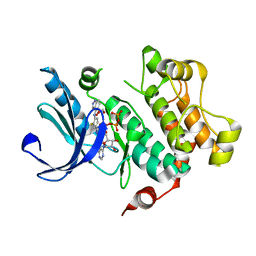 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | 4-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-1H-indazole-5-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
6GZ2
 
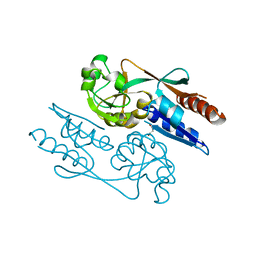 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, MALONATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
6GZ1
 
 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, SULFATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
8IY9
 
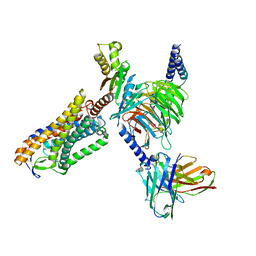 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYH
 
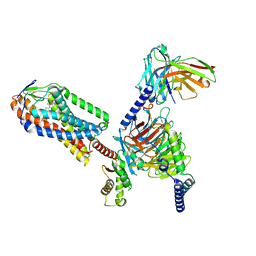 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYW
 
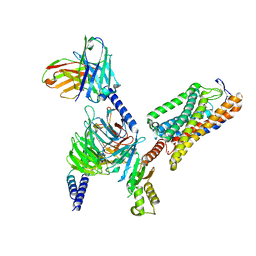 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JHN
 
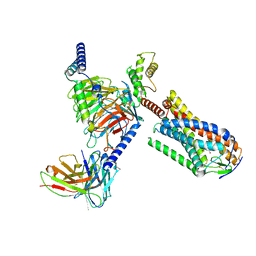 | | Structure of MMF-GPR109A-G protein complex | | Descriptor: | (E)-4-methoxy-4-oxidanylidene-but-2-enoic acid, G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
