2H6K
 
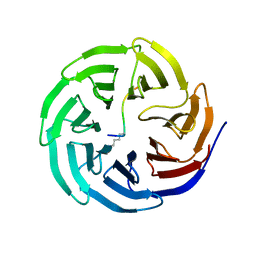 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6Q
 
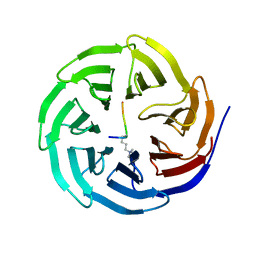 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me3 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GKU
 
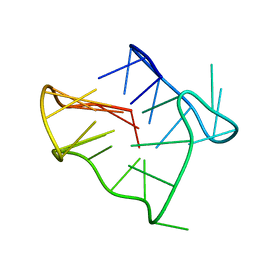 | | Monomeric human telomere DNA tetraplex with 3+1 strand fold topology, two edgewise loops and double-chain reversal loop, NMR, 12 structures | | Descriptor: | Human telomere DNA | | Authors: | Luu, K.N, Phan, A.T, Kuryavyi, V.V, Lacroix, L, Patel, D.J. | | Deposit date: | 2006-04-03 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in k(+) solution: an intramolecular (3 + 1) g-quadruplex scaffold.
J.Am.Chem.Soc., 128, 2006
|
|
6UF1
 
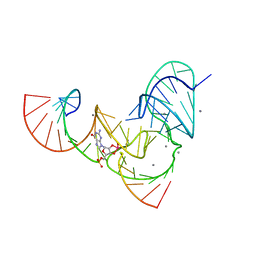 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UFJ
 
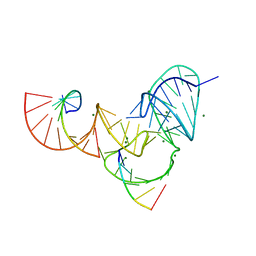 | | Pistol ribozyme product crystal structure | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UEY
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6V9Q
 
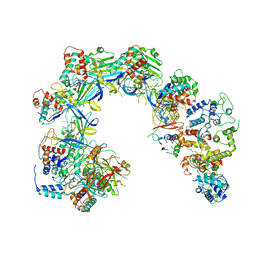 | | Cryo-EM structure of Cascade-TniQ binary complex | | Descriptor: | Cas8, RNA (61-MER), TniQ family protein, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VBW
 
 | |
6UFK
 
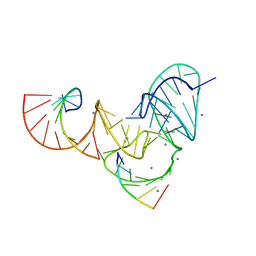 | | Pistol ribozyme product crystal soaked in Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6VRC
 
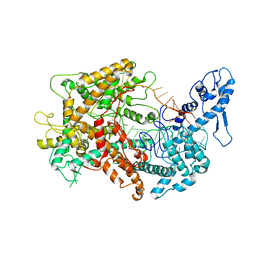 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
6V9P
 
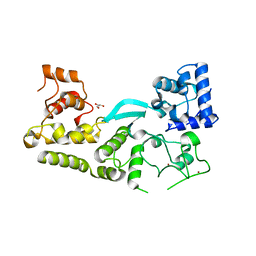 | | Crystal structure of the transposition protein TniQ | | Descriptor: | GLYCEROL, TniQ family protein, ZINC ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VRB
 
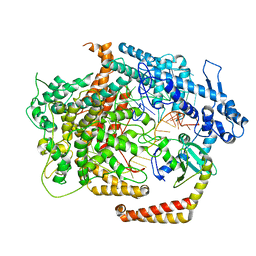 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
1EEG
 
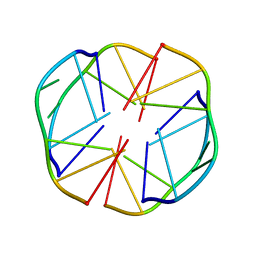 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
4FT2
 
 | |
1F3S
 
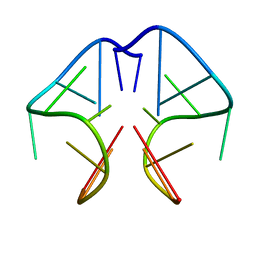 | | Solution Structure of DNA Sequence GGGTTCAGG Forms GGGG Tetrade and G(C-A) Triad. | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*TP*CP*AP*GP*G)-3') | | Authors: | Kettani, A, Basu, G, Gorin, A, Majumdar, A, Skripkin, E, Patel, D.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
J.Mol.Biol., 301, 2000
|
|
1FJB
 
 | | NMR Study of an 11-Mer DNA Duplex Containing 7,8-Dihydro-8-Oxoadenine (AOXO) Opposite Thymine | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(A38)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Chen, H, Johnson, F, Grollman, A.P, Patel, D.J. | | Deposit date: | 1995-12-15 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of the Ionizing Radiation Adduct 7,8-Dihydro-8-Oxoadenine (A Oxo) Positioned Opposite Thymine and Guanine in DNA Duplexes
MAGN.RESON.CHEM., 34, 1996
|
|
4HRE
 
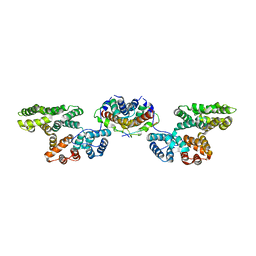 | | Crystal Structure of p11/Annexin A2 Heterotetramer in Complex with SMARCA3 Peptide | | Descriptor: | Annexin A2, Helicase-like transcription factor, Protein S100-A10 | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2012-10-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7852 Å) | | Cite: | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
4HRH
 
 | | Crystal Structure of p11-Annexin A2(N-terminal) Fusion Protein in Complex with SMARCA3 Peptide | | Descriptor: | Helicase-like transcription factor, Protein S100-A10, Annexin A2, ... | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2012-10-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
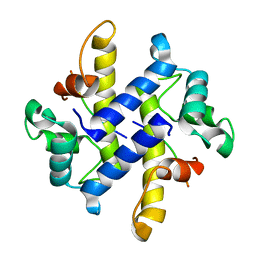
4HRG
 
 | |
4IUU
 
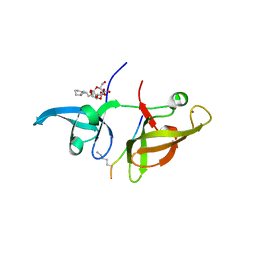 | |
4IUP
 
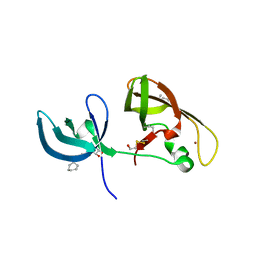 | |
1FMN
 
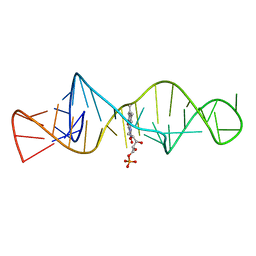 | | SOLUTION STRUCTURE OF FMN-RNA APTAMER COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (5'-R(*GP*GP*CP*GP*UP*GP*UP*AP*GP*GP *AP*UP*AP*UP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*AP*AP*GP *GP*AP*CP*AP*CP*GP*CP*C)-3') | | Authors: | Fan, P, Suri, A.K, Fiala, R, Live, D, Patel, D.J. | | Deposit date: | 1995-12-04 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the FMN-RNA aptamer complex.
J.Mol.Biol., 258, 1996
|
|
1FJA
 
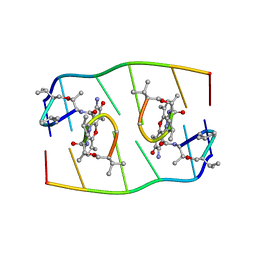 | |
4H9Q
 
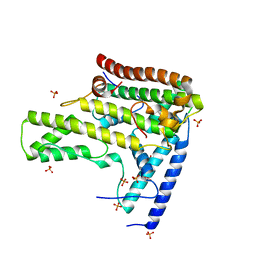 | | Complex structure 4 of DAXX(E225A)/H3.3(sub5)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
4IUQ
 
 | | crystal structure of SHH1 SAWADEE domain | | Descriptor: | CYMAL-4, SAWADEE HOMEODOMAIN HOMOLOG 1, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-01-21 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1.
Nature, 498, 2013
|
|
