6NJD
 
 | |
6NII
 
 | |
4M77
 
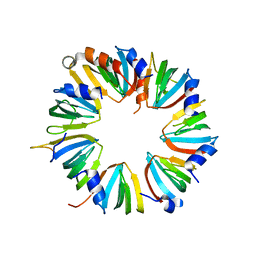 | | Crystal structure of Lsm2-8 complex, space group I212121 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M78
 
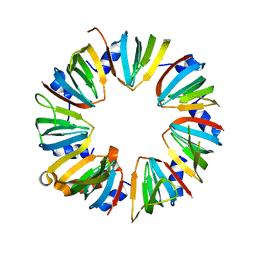 | | Crystal structure of Lsm2-8 complex, space group P21 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M7A
 
 | | Crystal structure of Lsm2-8 complex bound to the 3' end sequence of U6 snRNA | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M75
 
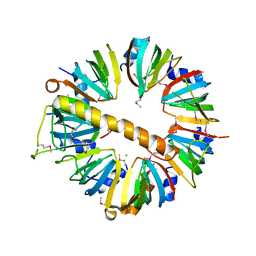 | | Crystal structure of Lsm1-7 complex | | Descriptor: | CHLORIDE ION, U6 snRNA-associated Sm-like protein Lsm1, U6 snRNA-associated Sm-like protein Lsm2, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4M7D
 
 | | Crystal structure of Lsm2-8 complex bound to the RNA fragment CGUUU | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
6WZT
 
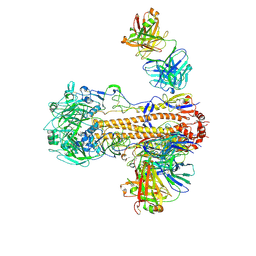 | | CryoEM structure of influenza hemagglutinin A/Victoria/361/2011 in complex with cyno antibody 3B10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cyno antibody heavy chain, ... | | Authors: | Qiu, Y, Zhou, Y, Darricarrere, N. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
6XDG
 
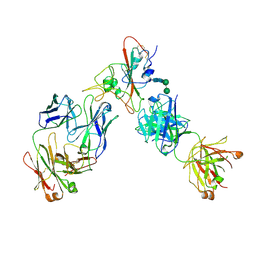 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
6XGC
 
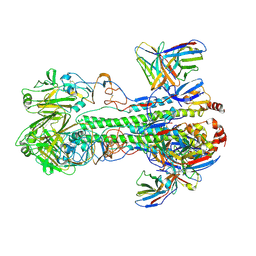 | |
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6LDU
 
 | |
6LE7
 
 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2 | | Descriptor: | (4R)-3-(2-hydroxyphenyl)-4-(3-methoxy-4-propoxy-phenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, Probable endochitinase | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85753441 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 2
To Be Published
|
|
6LE8
 
 | | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1 | | Descriptor: | (4R)-4-(4-ethoxyphenyl)-3-(2-hydroxyphenyl)-5-(pyridin-3-ylmethyl)-1,4-dihydropyrrolo[3,4-c]pyrazol-6-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chen, Q, Yang, Q, Zhou, Y. | | Deposit date: | 2019-11-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39909327 Å) | | Cite: | Crystal structure of nematode family I chitinase,CeCht1, in complex with dihydropyrrolopyrazol-6-one derivate 1
To Be Published
|
|
6MDR
 
 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
6PDX
 
 | |
8DTI
 
 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTH
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTF
 
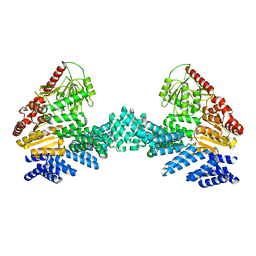 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
5JDB
 
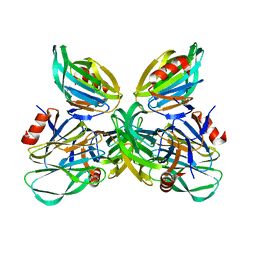 | | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens | | Descriptor: | Outer capsid protein VP4 | | Authors: | Sun, X, Guo, N, Li, D, Zhou, Y, Jin, M, Xie, G, Pang, L, Zhang, Q, Cao, Y, Duan, Z. | | Deposit date: | 2016-04-16 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Binding specificity of P[8] VP8* proteins of rotavirus vaccine strains with histo-blood group antigens.
Virology, 495, 2016
|
|
6XVD
 
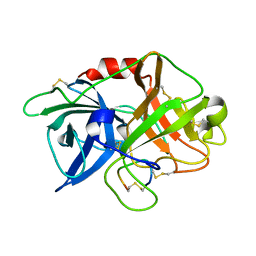 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1-W3F | | Authors: | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
8XEP
 
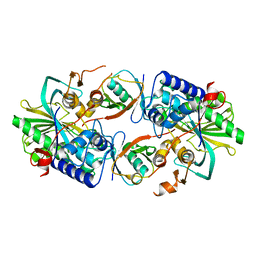 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | Descriptor: | SULFATE ION, Type IV effector MavL, Ubiquitin | | Authors: | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
4QXF
 
 | | crystal structure of human LGR4 and Rspo1 | | Descriptor: | Leucine-rich repeat-containing G-protein coupled receptor 4, Variable lymphocyte receptor B, R-spondin-1 | | Authors: | Xu, J.G, Huang, C, Zhou, Y, Zhu, Y. | | Deposit date: | 2014-07-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | crystal structure of human LGR4 and Rspo1
TO BE PUBLISHED
|
|
