4Y1B
 
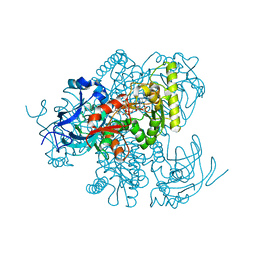 | |
9KM0
 
 | | Cryo-EM structure of a tri-heme cytochrome-associated RC-LH1 complex from a marine photoheterotrophic bacterium, purified with EDTA-2Na-containing solutions | | Descriptor: | (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate, (4~{E},16~{E},26~{E})-2-methoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-4,6,8,10,12,14,16,18,20,22,26,30-dodecaen-3-one, Antenna pigment protein alpha chain, ... | | Authors: | Chen, J.H. | | Deposit date: | 2024-11-15 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Cryo-EM Analysis of a Tri-Heme Cytochrome-Associated RC-LH1 Complex from the Marine Photoheterotrophic Bacterium Dinoroseobacter Shibae.
Adv Sci, 12, 2025
|
|
2NRN
 
 | |
7YT9
 
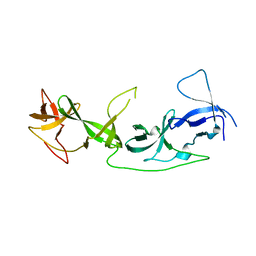 | | crystal structure of AGD1-4 of Arabidopsis AGDP3 | | Descriptor: | AGD1-4 of Arabidopsis AGDP3 | | Authors: | Zhou, X, Du, J. | | Deposit date: | 2022-08-13 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The H3K9me2-binding protein AGDP3 limits DNA methylation and transcriptional gene silencing in Arabidopsis.
J Integr Plant Biol, 64, 2022
|
|
7YTA
 
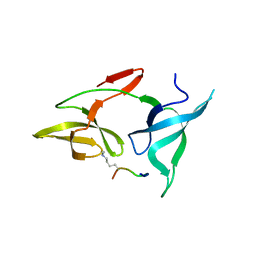 | |
8JTL
 
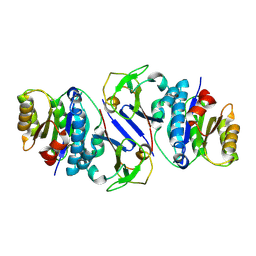 | | Structure of OY phytoplasma SAP05 binding with AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
8JTK
 
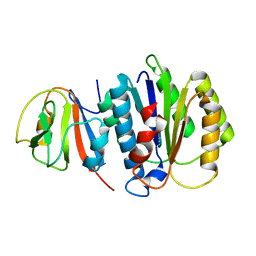 | | Structure of AYWB phytoplasma SAP05 recognizing AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
6LHC
 
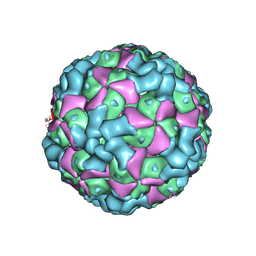 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
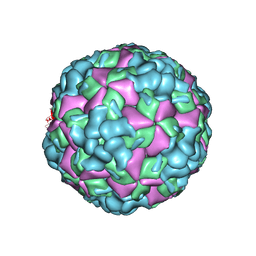 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
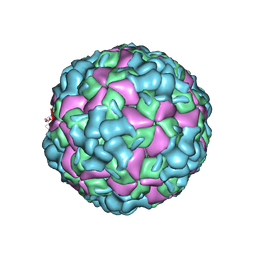 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
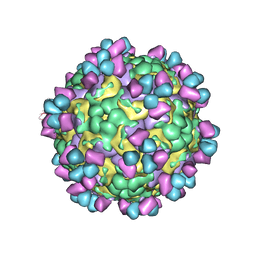 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHO
 
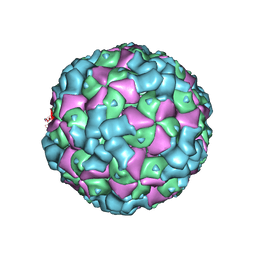 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHK
 
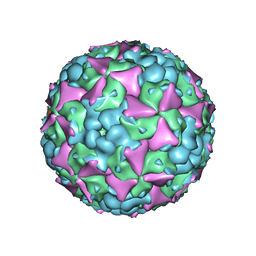 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
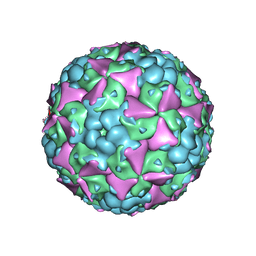 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
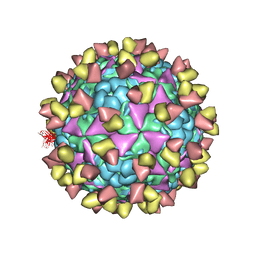 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
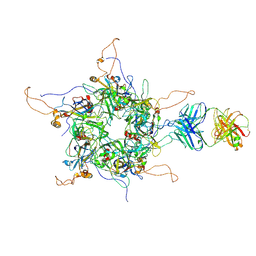 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
6LHT
 
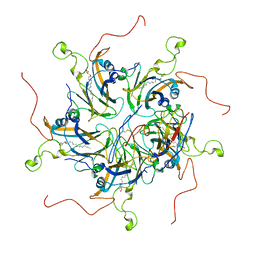 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
8GRM
 
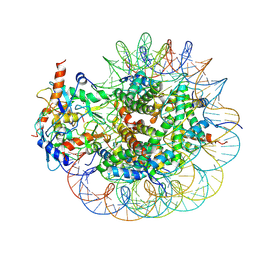 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GRQ
 
 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
6L31
 
 | | L1 protein of human papillomavirus 6 | | Descriptor: | Major capsid protein L1 | | Authors: | Li, S.W, Liu, X.L, Gu, Y. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Neutralization sites of human papillomavirus-6 relate to virus attachment and entry phase in viral infection.
Emerg Microbes Infect, 8, 2019
|
|
8KHL
 
 | | (S)-citramalyl-CoA lyase | | Descriptor: | COENZYME A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8KHG
 
 | | Itaconyl-CoA hydratase PaIch | | Descriptor: | CITRIC ACID, Itaconyl-CoA hydratase | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
7EAM
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
