6O8D
 
 | | Anti-CD28xCD3 CODV Fab bound to CD28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-CD28xCD3 CODV Fab Heavy chain, ... | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2019-03-09 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Trispecific antibodies enhance the therapeutic efficacy of tumor-directed T cells through T cell receptor co-stimulation
To Be Published
|
|
8STH
 
 | | human STING with diABZI agonist 15 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
8STI
 
 | | human STING with agonist XMT-1616 | | Descriptor: | 3-[(2E)-4-{5-carbamoyl-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-7-(3-hydroxypropoxy)-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(4-ethyl-2-methyl-1,3-oxazole-5-carbonyl)amino]-3H-imidazo[4,5-b]pyridine-6-carboxamide, Stimulator of interferon genes protein | | Authors: | Duvall, J.R, Bukhalid, R.A. | | Deposit date: | 2023-05-10 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Optimization of a STING Agonist Platform for Application in Antibody Drug Conjugates.
J.Med.Chem., 66, 2023
|
|
3N2N
 
 | | The Crystal Structure of Tumor Endothelial Marker 8 (TEM8) extracellular domain | | Descriptor: | ACETATE ION, Anthrax toxin receptor 1, MAGNESIUM ION | | Authors: | Fu, S, Tong, X.H, Wu, Y, Li, Y.Y, Rao, Z.H. | | Deposit date: | 2010-05-18 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of tumor endothelial marker 8 (TEM8) extracellular domain and implications for its receptor function for recognizing anthrax toxin.
Plos One, 5, 2010
|
|
8XGO
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGU
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
8XGS
 
 | | a peptide receptor complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Wu, Z, Du, Y, Chen, G. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for the ligand recognition and G protein subtype selectivity of kisspeptin receptor.
Sci Adv, 10, 2024
|
|
6AJ2
 
 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
7KEF
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
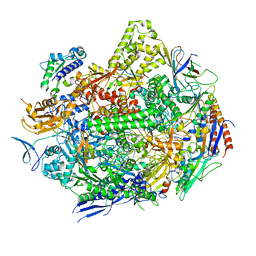 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
6AJ0
 
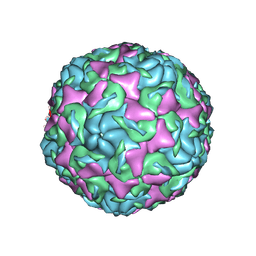 | | The structure of Enterovirus D68 mature virion | | Descriptor: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ9
 
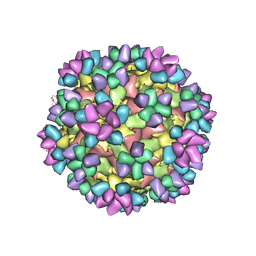 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ7
 
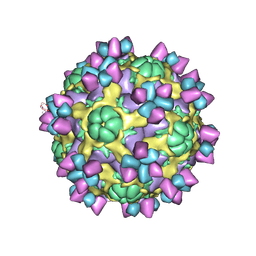 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
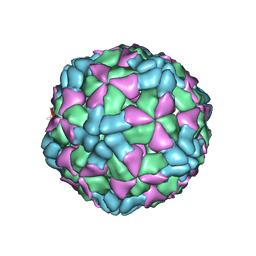 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
7XEM
 
 | | Cholesterol bound state of mTRPV2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
1OQD
 
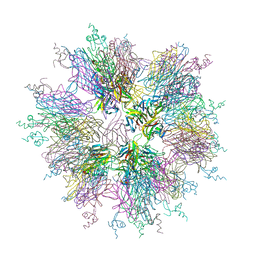 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
7XEU
 
 | | Structure of mTRPV2_E2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEO
 
 | | MbetaCD treated state of mTRPV2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEV
 
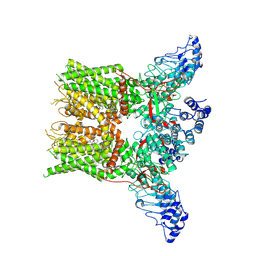 | | Structure of mTRPV2_2-APB | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEW
 
 | | Structure of mTRPV2_Q525F | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XER
 
 | | Structure of mTRPV2_Q525T | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
5DRB
 
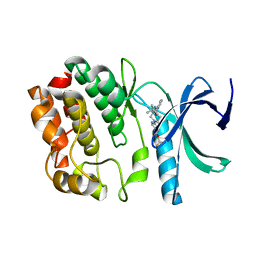 | | Crystal structure of WNK1 in complex with WNK463 | | Descriptor: | N-tert-butyl-1-(1-{5-[5-(trifluoromethyl)-1,3,4-oxadiazol-2-yl]pyridin-2-yl}piperidin-4-yl)-1H-imidazole-5-carboxamide, Serine/threonine-protein kinase WNK1 | | Authors: | Kohls, D, Xie, X. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Small-molecule WNK inhibition regulates cardiovascular and renal function.
Nat.Chem.Biol., 12, 2016
|
|
1OQE
 
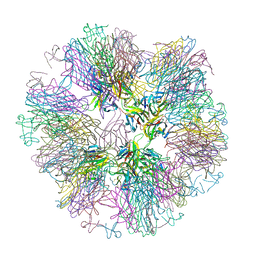 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
6LHB
 
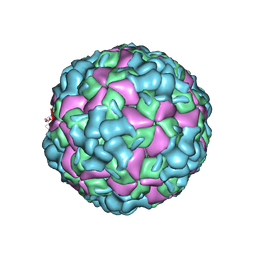 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
