2LX4
 
 | |
6U7E
 
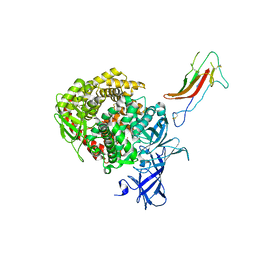 | | HCoV-229E RBD Class III in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7G
 
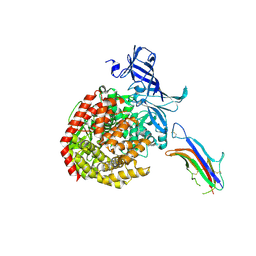 | | HCoV-229E RBD Class V in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
3B8N
 
 | |
3B8O
 
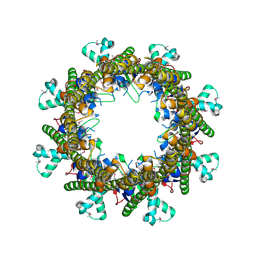 | |
3B8M
 
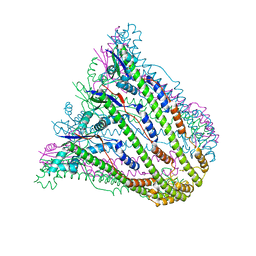 | |
3B8P
 
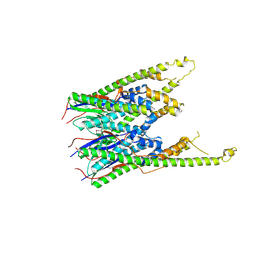 | |
3JVO
 
 | | Crystal structure of bacteriophage HK97 gp6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Gp6 | | Authors: | Lam, R, Tuite, A, Battaile, K.P, Edwards, A.M, Maxwell, K.L, Chirgadze, N.Y. | | Deposit date: | 2009-09-17 | | Release date: | 2009-11-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins.
J.Mol.Biol., 395, 2010
|
|
6OLJ
 
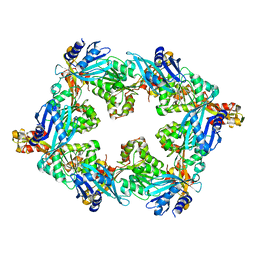 | |
5BXH
 
 | |
5BXD
 
 | |
6OK2
 
 | |
6OLL
 
 | |
8DNN
 
 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
6OJZ
 
 | |
5BXB
 
 | |
6OJY
 
 | |
6OLK
 
 | |
6OJX
 
 | |
6OKV
 
 | |
6OLM
 
 | |
5UFK
 
 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-04 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5UF5
 
 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila (domain-swapped dimer) | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VKK
 
 | |
5VOX
 
 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
