8KC1
 
 | | De novo design protein -NX5 | | Descriptor: | De novo design protein -NX5 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design protein -NX5
To Be Published
|
|
8KC8
 
 | | De novo design protein -T11 | | Descriptor: | De novo design protein -T11 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design protein -T11
To Be Published
|
|
8KA7
 
 | | De novo design protein -NB7 | | Descriptor: | De novo design protein -NB7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo design protein -NB7
To Be Published
|
|
8KA6
 
 | | De novo design protein -NA7 | | Descriptor: | De novo design protein -NA7 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo design protein -NA7
To Be Published
|
|
8KCJ
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein -N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo design protein -N7
To Be Published
|
|
8KC5
 
 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design protein -T09
To Be Published
|
|
8KC0
 
 | | De novo design protein -NB8 | | Descriptor: | De novo design protein -NB8 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design protein -NB8
To Be Published
|
|
8KCK
 
 | | De novo design protein -N9 | | Descriptor: | De novo design protein -N9 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | De novo design protein -N9
To Be Published
|
|
2MM5
 
 | |
2LYI
 
 | |
2MM6
 
 | |
2MAB
 
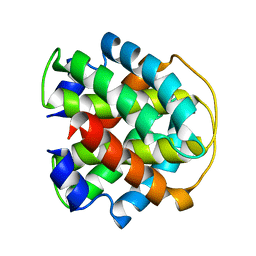 | |
7XUG
 
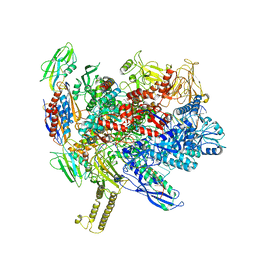 | |
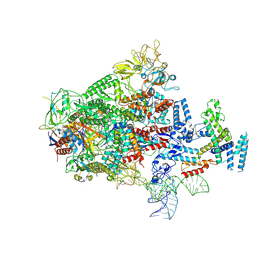
7XUI
 
 | |
7X5J
 
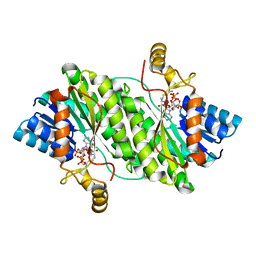 | | ACP-dependent oxoacyl reductase | | Descriptor: | 3-oxoacyl-ACP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLALANINE | | Authors: | Wang, S, Bai, L. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ACP-dependent oxoacyl reductase
To Be Published
|
|
7Y8T
 
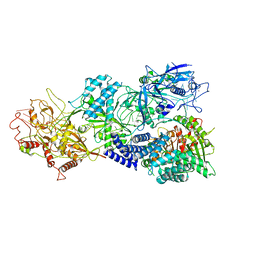 | | Structure of Cas7-11-crRNA in complex with TPR-CHAT | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (37-MER), ... | | Authors: | Wang, S, Guo, M, Zhu, Y, Huang, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the type III-E CRISPR-Cas effector gRAMP in complex with TPR-CHAT.
Cell Res., 32, 2022
|
|
7Y8Y
 
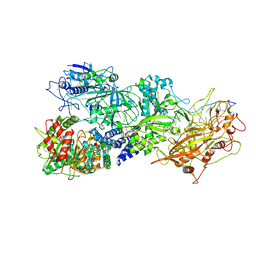 | | Structure of Cas7-11-crRNA-tgRNA in complex with TPR-CHAT | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (37-MER), ... | | Authors: | Wang, S, Guo, M, Zhu, Y, Huang, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the type III-E CRISPR-Cas effector gRAMP in complex with TPR-CHAT.
Cell Res., 32, 2022
|
|
4WKP
 
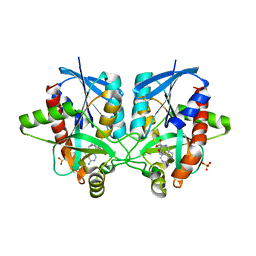 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 2-(2-hydroxyethoxy)ethylthiomethyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-(2-{[2-(2-hydroxyethoxy)ethyl]sulfanyl}ethyl)pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, SULFATE ION | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4BVJ
 
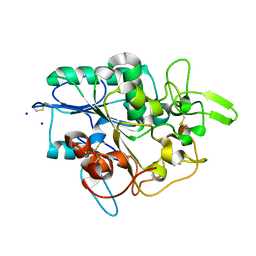 | | Structure of Y105A mutant of PhaZ7 PHB depolymerase | | Descriptor: | PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4WKN
 
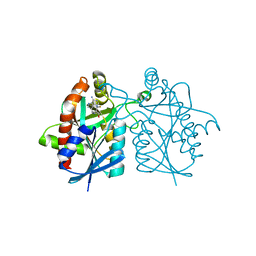 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKO
 
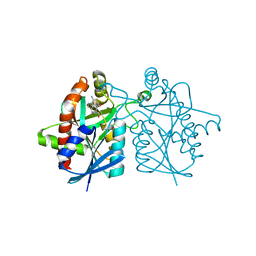 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | Authors: | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4KC4
 
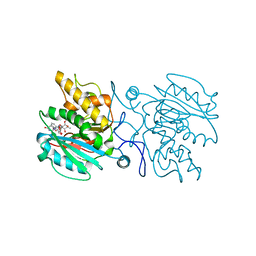 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 5'-deoxy-5'-[({6-[(alpha-D-galactopyranosyloxy)methyl]pyridin-2-yl}carbonyl)amino]uridine, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
4KC1
 
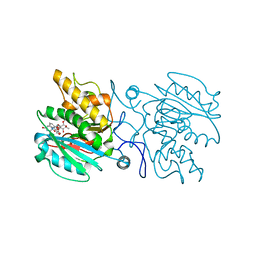 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 6-(1-beta-D-Glucopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
4KC2
 
 | | Structure of the blood group glycosyltransferase AAglyB in complex with a pyridine inhibitor as a neutral pyrophosphate surrogate | | Descriptor: | 6-(1-beta-D-Galactopyranosyloxymethyl)-N-(5'-deoxyluridine-5'-yl)picolinamide, Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase soluble form, MANGANESE (II) ION, ... | | Authors: | Cuesta-Seijo, J.A, Wang, S, Lafont, D, Vidal, S, Palcic, M.M. | | Deposit date: | 2013-04-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of glycosyltransferase inhibitors: pyridine as a pyrophosphate surrogate.
Chemistry, 19, 2013
|
|
7T7S
 
 | | R-27 in Complex with S. aureus DHFR and tricyclic-NADPH (tNADPH) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Reeve, S.M, Wang, S, Donald, B.R, Wright, D.L. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chiral evasion and stereospecific antifolate resistance in Staphylococcus aureus.
Plos Comput.Biol., 18, 2022
|
|
