3W9J
 
 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Multidrug resistance protein MexB, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | Authors: | Sakurai, K, Nakashima, R, Hayashi, K, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3W9H
 
 | | Structural basis for the inhibition of bacterial multidrug exporters | | Descriptor: | Acriflavine resistance protein B, [{2-[({[(3R)-1-{8-[(4-tert-butyl-1,3-thiazol-2-yl)carbamoyl]-4-oxo-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-4H-pyrido[1,2-a]pyrimidin-2-yl}piperidin-3-yl]oxy}carbonyl)amino]ethyl}(dimethyl)ammonio]acetate | | Authors: | Sakurai, K, Nagata, C, Nakashima, R, Yamaguchi, A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the inhibition of bacterial multidrug exporters
Nature, 500, 2013
|
|
3VYW
 
 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
3W1W
 
 | | Protein-drug complex | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHOLIC ACID, ... | | Authors: | Ishii, R, Gupta, V, Yamaguchi, Y, Handa, H, Nureki, O. | | Deposit date: | 2012-11-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Salicylic Acid induces mitochondrial injury by inhibiting ferrochelatase heme biosynthesis activity
Mol.Pharmacol., 84, 2013
|
|
5JMF
 
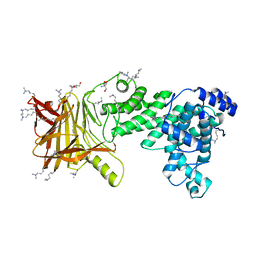 | | Heparinase III-BT4657 gene product | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Heparinase III protein, ... | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
3O39
 
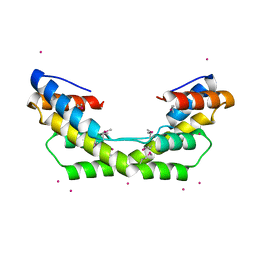 | |
4X8Q
 
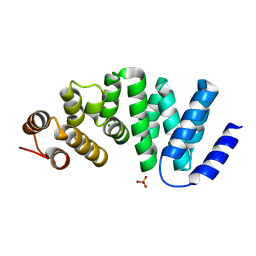 | | X-ray crystal structure of AlkD2 from Streptococcus mutans | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mullins, E.A, Shi, R, Eichman, B.F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A New Family of HEAT-Like Repeat Proteins Lacking a Critical Substrate Recognition Motif Present in Related DNA Glycosylases.
Plos One, 10, 2015
|
|
3OQ5
 
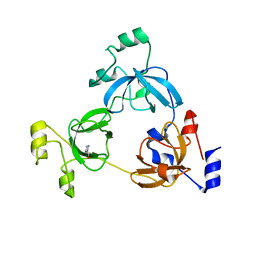 | | Crystal structure of the 3-MBT domain from human L3MBTL1 in complex with p53K382me1 | | Descriptor: | Cellular tumor antigen p53, Lethal(3)malignant brain tumor-like protein | | Authors: | Roy, S, West, L.E, Weiner, K.L, Hayashi, R, Shi, X, Appella, E, Gozani, O, Kutateladze, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5005 Å) | | Cite: | The MBT Repeats of L3MBTL1 Link SET8-mediated p53 Methylation at Lysine 382 to Target Gene Repression.
J.Biol.Chem., 285, 2010
|
|
8D8Z
 
 | | Crystal structure of ChoE N147A mutant in complex with thiocholine and chloride | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, CHLORIDE ION, ChoE, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D91
 
 | | Crystal structure of ChoE in complex with acetate and tetraethylammonium (TEA) | | Descriptor: | ACETATE ION, ChoE, TETRAETHYLAMMONIUM ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8W
 
 | | Crystal structure of ChoE with Ser38 adopting alternative conformations | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ChoE, IODIDE ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8Y
 
 | | Crystal structure of ChoE N147A mutant in complex with acetylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETYLTHIOCHOLINE, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D90
 
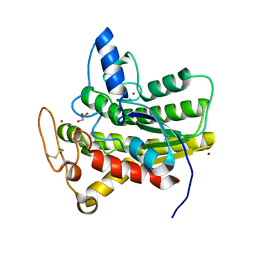 | | Crystal structure of ChoE N147A mutant in complex with bromide ions | | Descriptor: | BROMIDE ION, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8X
 
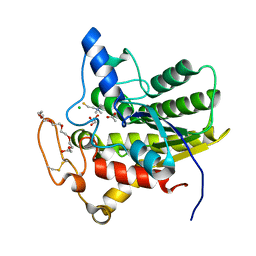 | | Crystal structure of ChoE in complex with acetate and thiocholine (crystal form 2) | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8TW1
 
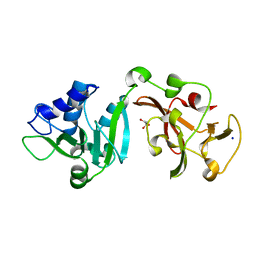 | | Crystal structure of Lys2972, a phage endolysin targeting Streptococcus thermophilus | | Descriptor: | Endolysin Lys2972, GLYCEROL, SODIUM ION | | Authors: | Zhu, X, Moineau, S, Shi, R. | | Deposit date: | 2023-08-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Fermentation Practices Select for Thermostable Endolysins in Phages.
Mol.Biol.Evol., 41, 2024
|
|
5JMD
 
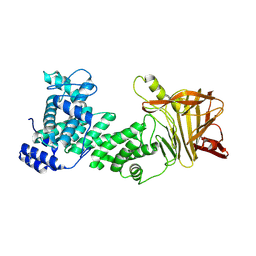 | | Heparinase III-BT4657 gene product, Methylated Lysines | | Descriptor: | Heparinase III protein, MAGNESIUM ION | | Authors: | Ulaganathan, T.S, Shi, R, Yao, D, Garron, M.-L, Cherney, M, Cygler, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-05-25 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility of PL12 family heparinases: structure and substrate specificity of heparinase III from Bacteroides thetaiotaomicron (BT4657).
Glycobiology, 27, 2017
|
|
6UQW
 
 | |
6UR1
 
 | |
6UQZ
 
 | |
6UQY
 
 | |
6UR0
 
 | | Crystal structure of ChoE D285N mutant acyl-enzyme | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
6UQX
 
 | | Crystal structure of ChoE in complex with propionylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ChoE, IODIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
8VLK
 
 | | Crystal structure of the yeast cytosine deaminase containing both open and closed active sites | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, SULFATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLL
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VLJ
 
 | | Crystal structure of the cacodylate-bound yeast cytosine deaminase (closed form) | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, Cytosine deaminase, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
