4QX0
 
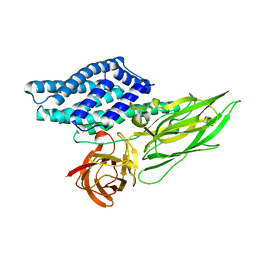 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX1
 
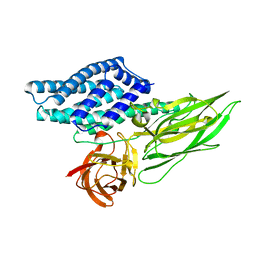 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2CLI
 
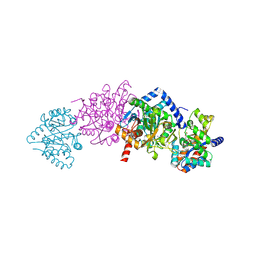 | | Tryptophan Synthase in complex with N-(4'- trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
2FE6
 
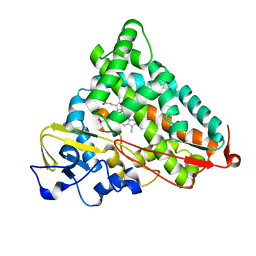 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | Cytochrome P450-cam, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING MN | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
2FXM
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | MERCURY (II) ION, Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FXO
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FEU
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
2FER
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | Cytochrome P450-cam, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING MN | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
2CLH
 
 | | Tryptophan Synthase in complex with (naphthalene-2'-sulfonyl)-2-amino- 1-ethylphosphate (F19) | | Descriptor: | 2-[(2-NAPHTHYLSULFONYL)AMINO]ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex.
Biochemistry, 46, 2007
|
|
2CLF
 
 | | Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2- amino-1-ethylphosphate (F6) - highF6 complex | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
2CLK
 
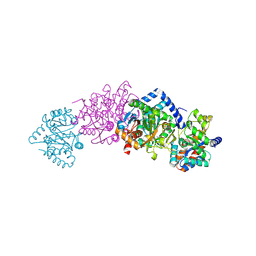 | | Tryptophan Synthase in complex with D-glyceraldehyde 3-phosphate (G3P) | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
4HU4
 
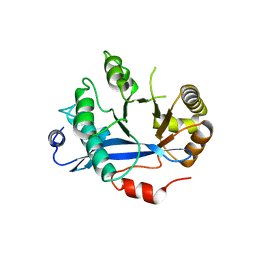 | | Crystal structure of EAL domain of the E. coli DosP - dimeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HSE
 
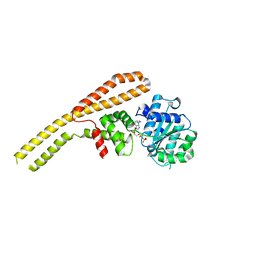 | | Crystal structure of ClpB NBD1 in complex with guanidinium chloride and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Chaperone protein ClpB, ... | | Authors: | Zeymer, C, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular mechanism of hsp100 chaperone inhibition by the prion curing agent guanidinium chloride.
J.Biol.Chem., 288, 2013
|
|
1LGF
 
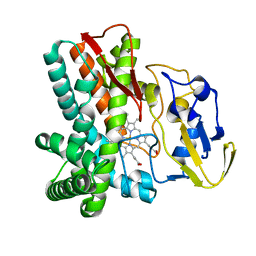 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
1LG9
 
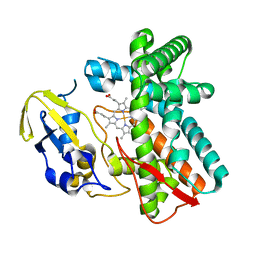 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-15 | | Release date: | 2002-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
4J80
 
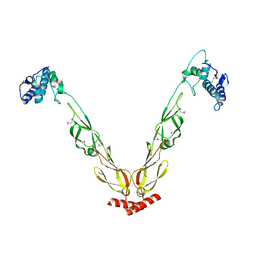 | | Thermus thermophilus DnaJ | | Descriptor: | Chaperone protein DnaJ 2 | | Authors: | Barends, T.R.M, Brosi, R.W, Steinmetz, A, Scherer, A, Hartmann, E, Eschenbach, J, Lorenz, T, Seidel, R, Shoeman, R, Zimmermann, S, Bittl, R, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | EPR (2.9 Å), X-RAY DIFFRACTION | | Cite: | Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J7Z
 
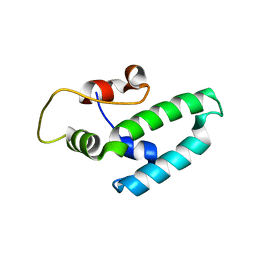 | | Thermus thermophilus DNAJ J- and G/F-DOMAINS | | Descriptor: | Chaperone protein DnaJ 2, GLYCEROL | | Authors: | Barends, T.R.M, Brosi, R.W, Steinmetz, A, Scherer, A, Hartmann, E, Eschenbach, J, Lorenz, T, Seidel, R, Shoeman, R, Zimmermann, S, Bittl, R, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | EPR (1.64 Å), X-RAY DIFFRACTION | | Cite: | Combining crystallography and EPR: crystal and solution structures of the multidomain cochaperone DnaJ.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1N9L
 
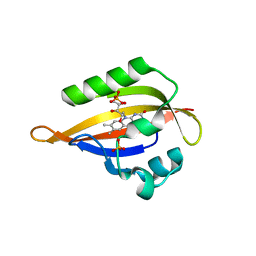 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in the dark state. | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
1N9O
 
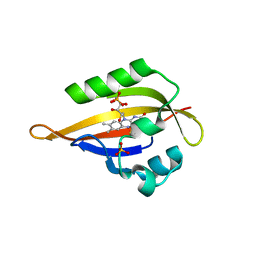 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Composite data set. | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
4HH1
 
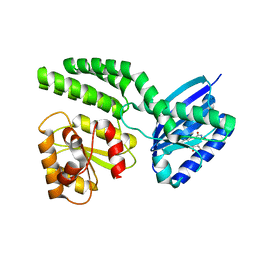 | | Dark-state structure of AppA wild-type without the Cys-rich region from Rb. sphaeroides | | Descriptor: | AppA protein, FLAVIN MONONUCLEOTIDE | | Authors: | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | Deposit date: | 2012-10-09 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1N9N
 
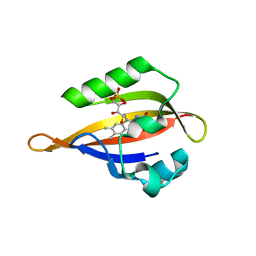 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Data set of a single crystal. | | Descriptor: | FLAVIN MONONUCLEOTIDE, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
1LFK
 
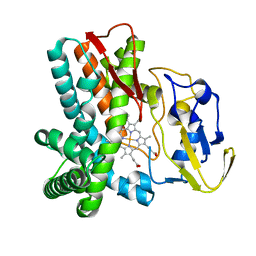 | | Crystal structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction During Vancomycin Biosynthesis | | Descriptor: | P450 monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pylypenko, O, Zerbe, K, Vitali, F, Zhang, W, Vrijbloed, J.W, Robinson, J.A, Schlichting, I. | | Deposit date: | 2002-04-11 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of OxyB, a Cytochrome P450 Implicated in an Oxidative Phenol Coupling Reaction during Vancomycin Biosynthesis.
J.Biol.Chem., 277, 2002
|
|
4K9R
 
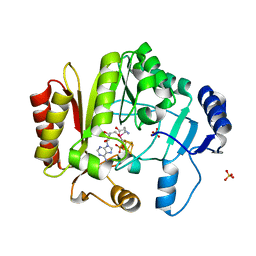 | | Spore photoproduct lyase Y98F mutant | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Spore photoproduct lyase, ... | | Authors: | Yang, L, Nelson, R.S, Benjdia, A, Lin, G, Telser, J, Stoll, S, Schlichting, I, Li, L. | | Deposit date: | 2013-04-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A radical transfer pathway in spore photoproduct lyase.
Biochemistry, 52, 2013
|
|
4HU3
 
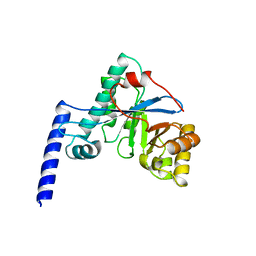 | | Crystal structure of EAL domain of the E. coli DosP - monomeric form | | Descriptor: | Oxygen sensor protein DosP | | Authors: | Tarnawski, M, Barends, T.R.M, Hartmann, E, Schlichting, I. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structures of the catalytic EAL domain of the Escherichia coli direct oxygen sensor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LJ5
 
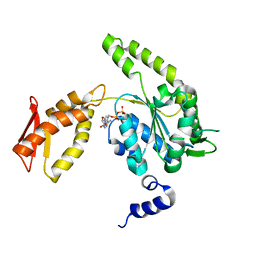 | | ClpB NBD2 from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
