2OEW
 
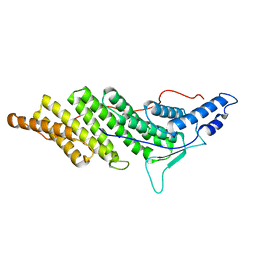 | | Structure of ALIX/AIP1 Bro1 Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2OEV
 
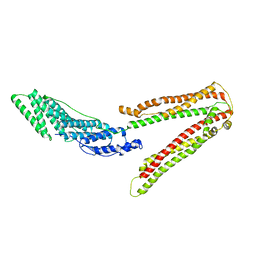 | | Crystal structure of ALIX/AIP1 | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2R0V
 
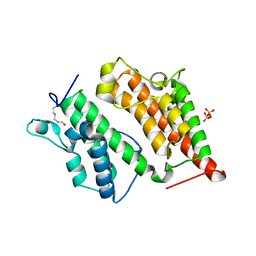 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0Y
 
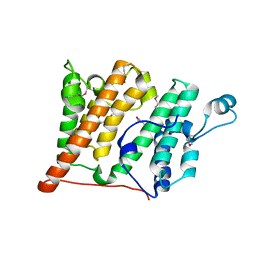 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0S
 
 | | Crystal Structure of the Rsc4 tandem bromodomain | | Descriptor: | Chromatin structure-remodeling complex protein RSC4 | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R10
 
 | | Structure of an acetylated Rsc4 tandem bromodomain Histone Chimera | | Descriptor: | 1,2-ETHANEDIOL, Chromatin structure-remodeling complex protein RSC4, LINKER, ... | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R3C
 
 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | Descriptor: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7JX4
 
 | | Crystal Structure of N-Lysine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Lysine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices.
J.Am.Chem.Soc., 143, 2021
|
|
7JX5
 
 | | Crystal Structure of N-Phenylalanine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Phenylalanine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices
J.Am.Chem.Soc., 143, 2021
|
|
3D8T
 
 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | ACETATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8S
 
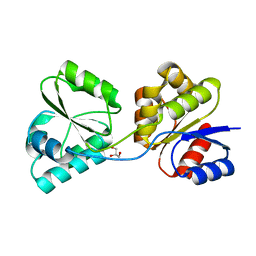 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | GLYCEROL, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8N
 
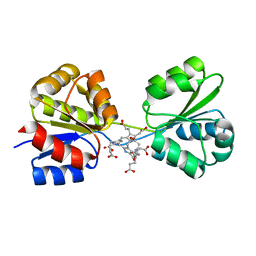 | | Uroporphyrinogen III Synthase-Uroporphyringen III Complex | | Descriptor: | 3,3',3'',3'''-[3,8,13,17-tetrakis(carboxymethyl)porphyrin-2,7,12,18-tetrayl]tetrapropanoic acid, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
3D8R
 
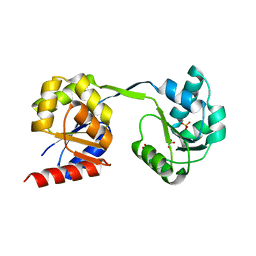 | | Thermus thermophilus Uroporphyrinogen III Synthase | | Descriptor: | PHOSPHATE ION, Uroporphyrinogen-III synthase | | Authors: | Schubert, H.L. | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
Biochemistry, 47, 2008
|
|
1PNL
 
 | |
1GM7
 
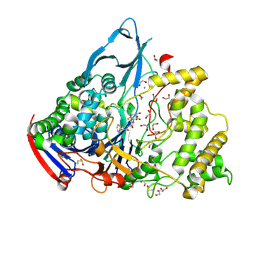 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM9
 
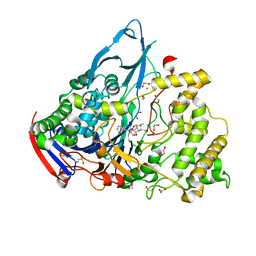 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-12 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1PNK
 
 | |
1PNM
 
 | |
1GM8
 
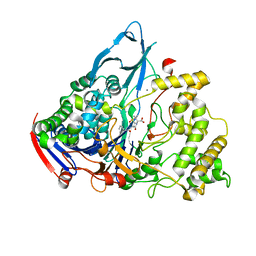 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-11 | | Release date: | 2001-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GKF
 
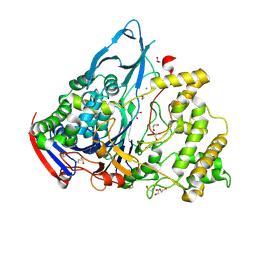 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-13 | | Release date: | 2002-01-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GMP
 
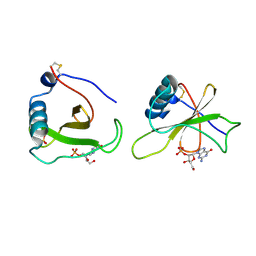 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMR
 
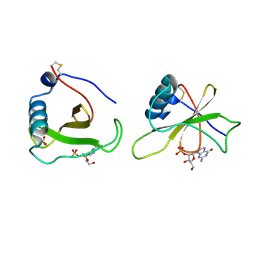 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMQ
 
 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
